Introduction
The potato, a tuberous herbaceous plant belonging to the Solaneaceae family, is one of the most important crops worldwide, occupying the third place in consumption. In Peru, 19 out of 24 regions practice potato agriculture (Fonseca et al. 2014); and it is one of the main sources of food and income for low-income citizens. Therefore, improving the yield and phytosanitary care during their cultivation is essential for food security (Chávez 2008).
Globodera pallida (Stone) a white potato cyst nematode is widely distributed in the Peruvian Andean, being responsible for significant economic losses in potato yield (Evans et al. 1975, Eves-van den Akker et al. 2016, Jones et al. 2013, Franco et al. 1990). Cyst nematodes are parasites that penetrate the root and migrate to the intracellular spaces of the host plant. They induce the fusion of approximately 200 neighboring protoplasts, in the sedentary phase, after the partial dissolution of the cell walls. Thus, resulting in multinucleated giant hypertrophic cells called a syncytium (Kindt et al. 2013).
During the development of the nematodes, these syncytia continuously increase in volume by incorporating neighboring cells, through the rupture of their cell walls. Syncytia is a source of nutrients for the nematode, where the female is permanently fixed (Sobczak et al. 2011). Once the female is fertilized, it produces eggs that remain inside her body. Subsequently, the female becomes a cyst that houses these viable eggs, which protects against possible adverse environmental conditions, for up to more than 30 years (Curtis 2007).
Plant resistance to Globodera sp. would involve the expression of genes associated with transcription regulation, resistance signaling pathways, the defense mechanism and cell cycle regulation (Jolivet et al. 2007). Also, genes specialized in the recognition of specific pathogenic effectors generated by the causal agent of the disease would be involved. These specific recognition genes are called R genes. Most encode proteins with functional domains: NBS (nucleotide binding site) and LRR: (Leucine rich repeats). The last one determines the specificity for the pathogen recognition (Williamson 1999).
Among the R genes against G. pallida identified in potato, Gpa2 is the first nematode specific R gene to have been cloned. It has the characteristic of dominance (Van der Vossen et al. 2000). On the other hand, two QTLs associated to G. pallida resistance, Gpa VsspI and Gpa Xlsspl, located on chromosome 5 and 11, respectively, have also been identified in potato. It was observed that the presence of both QTLs in the plant generated a necrotic reaction in the roots of infected potatoes, which in turn led to a considerable reduction in the population of females (Caromel et al. 2005). In an analysis of the coding sequences of the Solanum tuberosum group Phureja genome, a susceptible variety, 435 NBS-encoding R gene homologs and 142 NBS-derived genes were annotate. Of the NBS-derived genes, 41% were pseudo-genes (Lozano et al. 2012).
The identification of nematode resistance genes is essential for the study of plant-pathogen interactions at the molecular and biochemical level. Then, it is necessary to carry out new exhaustive investigations to identify resistance genes against Globodera.
One of the more recent methods for identifying new genes is RNA Sequencing (RNA-seq) or massive transcriptome sequencing (Kochetov 2017). RNA-Seq has become instrumental to the discovery of new genes involved in resistance to various pathogens in plants (Wang et al 2016, Kong et al 2015, Wang et al. 2015). However, a global analysis of the potato transcriptome, at the moment Globodera pallida infection, has yet to be carried out.
In this study, we performed an RNA-seq to identify differentially expressed genes between two potato varieties, one resistant and another susceptible to G. pallida, under conditions of nematode infection. This differential expression at the gene level allowed us to identify candidate genes linked to resistance to G. pallida.
Material and methods
Biological material. The improved variety “María Huanca” (CIP code 279142.12), was used as the resistant genotype, comes from the cross between Solanum andigena (CIP code 276012.24) and AM66-246, the latter resulting from a backcross of Solanum vernei with S. tuberosum subsp. tuberosum (Llontop et al. 1989). The susceptible genotype used was the native variety “Chimbina Colorada” (Solanum chaucha, Code CIP 701013), resulting from the natural hybridization between Solanum tuberosum (2n = 4x = 48) and the diploid species Solanum stenotomum (2n = 2x = 24).
The Globodera pallida cysts were extracted from soil samples collected in infested areas of the province of Cusco, Peru. The Fenwick’s modified apparatus (Fenwick 1940) was used to isolate the cysts from the soil samples. It has a funnel through which the soil sample passes (20 meshes/cm2 or 850 μm of opening), falling into a container with water. The floating material is filtered by washing with water jets through the sieve of 60 mesh/cm2 (250 μm) where organic matter is retained, followed washing by another sieve of 120 mesh/cm2 (125 μm) where Globodera cysts are retained.
The isolated cysts were disinfected with 90% ethanol, 1.3% bleach and sterile distilled water (Heungens et al. 1996). The identification of the species was done using specific primers for the detection and amplification of ITS region (intergenic space) that distinguishes the two species of the genus Globodera sp. by means of the molecular size of the amplified bands. The expected band size for G. pallida is 434 bp (White et al. 1990, Bulman et al.,1997).
Experiment Design. We used 3-week old, aeroponically grown clonal potato plants of the “Maria Huanca” and “Chimbina Colorada”. The aeroponic system ensured the sustainable and abundant growth of the root system, of most importance during the infection stage with Globodera pallida.
To obtain the infective stage of juvenile 2 (J2) G. pallida, sets of 20 isolated and disinfected nematode cysts were immersed in distilled water for 3 days and further they were submerged in root exudate from potato plants susceptible variety “Yungay”, modified by Franco (1990). To filter the root exudate, we used Whatman No. 1 filter paper and a sterile Chromafil®Xtra PES-20/25 filter of 0.20 μm pore size. Each set of cysts was placed on a watch glass. The system was maintained at a temperature between 12-15 °C. Every 3 days, observations of the J2 emergence of G. pallida were made, using Carl Zeiss Stemi™DV4 stereoscope. Correspondingly, the root exudate was renewed to avoid the presence of fungi or polluting agents in the system. For the infection tests, the root system of the 3-week-old seedlings was carefully placed in the aeroponic system in 150 x 15 mm petri dishes containing 2% water-agar (Franco 1990) (Figure 1).
The experimental design for each variety of potato (“María Huanca” and “Chimbina Colorada”) consisted of a control treatment (without inoculation of G. pallida J2) and another treatment with infection of G. pallida J2 at 72 hours post inoculation (hpi), each treatment with 3 repetitions (Table 1). In each experimental unit (plant), five inoculation points were determined at the level of the root elongation zone, at each point 20 J2 of G. pallida were inoculated (Figure 1).
Differential expression analysis. The total RNA of 1-2 g of root of the resistant and susceptible plants was extracted from the treatments and controls after 72 hpi, using Tri®Reagent (Sigma). To remove the remaining DNA, the samples were treated with DNA-freeTM Kit (Ambion, USA), following the manufacturer's instructions. These samples were sent for sequencing to the company Clinical Microarray Core UCLA (Los Angeles, United States), where the verification of the samples quality, the construction of the libraries and the massive sequencing of the RNA was carried out, using the Illumina platform HiSeq-2500. A total of twelve samples were sequenced, as shown in Table 1. For the analysis of genetic expression of the two potato varieties with respect to the invasion of juveniles 2 (J2) of G. pallida, we used the bioinformatics platform installed on the server of the Genomics Research Unit-UPCH. The quality control of the sequencing read was performed using FASTQC, using a threshold between 28 and 40. Sequences with lower scores were discarded using FAST_X. We mapped the reads to the potato reference genome (PGSC 2011) corresponding to S. tuberosum group Phureja DM1-3 516R44 v.4.03, using bowtie2 v2.2.3 (Langmead & Salzberg 2012) and TopHat v2.0.12 (Kim et al. 2013). The alignments generated by TopHat were used as an input file for the Cufflinks v2.2.1 (Trapnell 2010) to determine the differential expression of genes, based on normalized levels of gene expression; that is, reads per kilo base per million mapped read (RPKM).

Figure 1 Experimental unit. Potato Plant of 3 weeks of growth in aeroponic system on 2% water agar support. Circles indicate the elongation zones of roots where Globodera pallida J2 were inoculated.
Table 1 Description of 12 libraries. S: susceptible, R: resistant.
| Variety | Treatment | Repetition | Libraries | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Susceptible | 0 hpi (control) | 1 | S1 | 2 | S2 | 3 | S3 | 72 hpi (infection) | 1 | S1 | 2 | S2 | 3 | S3 |
| Resistant | 0 hpi (control) | 1 | R1 | 2 | R2 | 3 | R3 | 72 hpi (infection) | 1 | R4 | 2 | R5 | 3 | R6 |
Then, when comparing the values of RPKM recorded in the control treatments (without inoculation of J2) and the 72 hpi infection treatment with J2, changes were observed in the expression levels of the mapped genes, concluding that the potato varieties modulated their expression against the juveniles 2 (J2) of Globodera pallida.
The list of genes obtained as a result of the previous analysis was subjected to inclusion criteria, obtaining a new select group of genes that demonstrated significant expression regulation. The criterion applied were: a) Genes that were significantly expressed, p <0.05; b) Fold Change ≥2 genes was deemed as overexpression; and Fold change ≤ -2 underexpression. We selected the R genes that were in the range -1 ≤ Fold Change ≤ 1; Finally, c) the opposite behavior at the gene expression level: over-expression and underexpression or invested, in resistant and susceptible varieties.
Results and discussion
Quality control. As a result of RNA-seq, 12 libraries of between 26-36 million reads were generated (Table 2). Using FASTQC, regions with low quality (Phred score <28) were discarted to retain only trust sequences for next subsequent analyzes.
Mapping of reads. Using TopHat, sequences that did not align to the DM potato reference genome were discarded, retaining only the aligned readings. The total number of aligned reads within libraries was in a percentage range of 76.7% - 91.4% (Table 2).
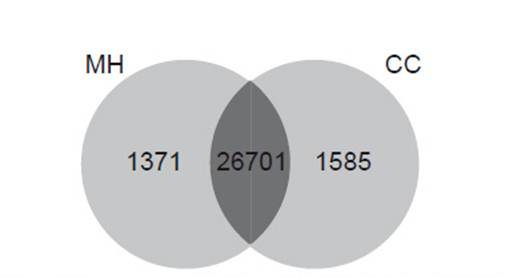
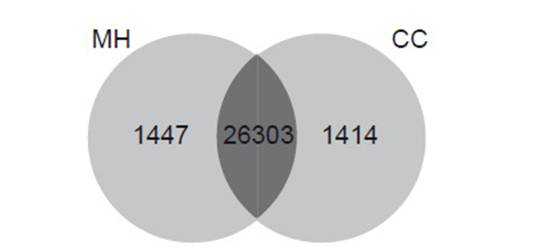
Differential gene expression. Using Cufflinks, we found that the values of RPKM showed variation in the number of genes expressed in treatments. In control treatment (0 hpi), a total of 1371 genes were uniquely expressed in “Maria Huanca” (variety resistant to Globodera pallida), and a total of 1585 were uniquely expressed in “Chimbina Colorada” (susceptible). 26701 genes were expressed in both varieties, in control treatment (0 hpi) (Fig. 2). However, under the infected treatment (72 hpi), a total of 1447 and 1414 uniquely expressed genes induced in resistant and susceptible variety were found, respectively; and 26303 genes were expressed in both varieties (Fig. 3).
Table 2 Quantity and percentage of reads mapped within the genome of DM1-3 516R44. The table shows the total number of reads resulting from sequencing (total sequences), and how many of these reads came to align to potato reference genome, using TopHat (mapped sequences), together with its corresponding percentage value (% of mapped sequences).
| Libraries | Total Sequences | Mapped Sequences | Percentage of Mapped Sequences |
|---|---|---|---|
| S1.fq | 31944894 | 29106799 | 91.10% |
| S2.fq | 32233705 | 29327225 | 91% |
| S3.fq | 32763288 | 29961755 | 91.40% |
| S4.fq | 31918298 | 27399347 | 85.80% |
| S5.fq | 34115485 | 29944518 | 87.80% |
| S6.fq | 36291159 | 27844552 | 76.70% |
| R1.fq | 29038036 | 24861409 | 85.60% |
| R2.fq | 28665423 | 24995899 | 87.20% |
| R3.fq | 26947639 | 22776382 | 84.50% |
| R4.fq | 29420533 | 23195366 | 78.80% |
| R5.fq | 32652723 | 25939737 | 79.40% |
| R6.fq | 30080914 | 24541192 | 81.60% |
Comparing values of RPKM and Fold Change (FC), we found a group of 100 genes with significant differences in gene expression (p <0.05) between the control (0 hpi) and infection (72 hpi) treatments. In “Maria Huanca” (resistant variety), we found 2113 and 3088 over and underexpressed genes, respectively. With respect to “Chimbina Colorada” (susceptible variety), we found 3561 and 3440 over and under expressed genes, respectively (Figure 4).
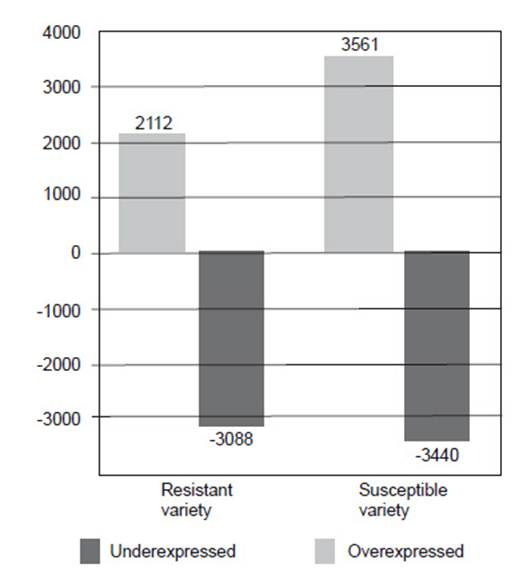
Figure 4 Number of genes that were overexpressed and underexpressed significantly (p <0.05) in the resistant variety (María Huanca) and susceptible (Chimbina Colorada), comparing control (0 hpi) and infection (72 hpi) treatments.
Within these groups, UDP-glucosyltransferase (locus ST4.03ch10: 4495601-4497176) behaved in an opposite manner between the resistant and susceptible varieties (Table 3). This gene is related to carbohydrate biosynthesis and cell wall synthesis (Schaff et al. 2007) and was observed in the resistant variety “Maria Huanca” at 72 hpi with juveniles 2 of Globodera pallida. According to the interaction between Solanum torvum - Meloidogyne incognita and Boehmeria nívea - Pratylenchus coffeae, a high concentration of UDP-glucosyltransferases in the plant confers resistance to the pathogen (Bagnaresi et al. 2013 and Yu et al. 2015). Likewise, the increase in the expression of LRX leucine-rich repeat extension gene (locus ST4.03ch11: 2710622-2713343) was observed. This gene encodes an extracellular protein that is bound to the cell wall, which plays an essential role in the formation of the cell wall and defense against pathogens (Draeger et al. 2015). LRX1 has been characterized as a member of the extracellular protein family LRR/extension. This gene is an important factor in the process of root hair development, which suggests that LRX1 represents a component of the cell wall involved in cell expansion (Baumberger et al. 2001). The inverse behavior of both genes was found in the susceptible variety.
Table 3 Genes in common found in both potato varieties, which overexpressed and/or underexpressed after 72 hours post inoculation. The change at the level of gene expression was measured based on the fold change values, which is the log2 (RPKM post infection/RPKM control). A positive fold change value implies overexpressed, while a negative value implies underexpressed. No INF: Not infected (0 hpi), INF: Infected (72 hpi)
| Genes | María Huanca | Chimbina Colorada | |||||
|---|---|---|---|---|---|---|---|
| Name | No INF | INF | Fold Change | No INF | INF | Fold Change | |
| COL domain class transcription factor | 0.44 | 2.25 | 2.36 | 0.74 | 0.33 | -1.18 | |
| UDP-glucosyltransferase | 0.32 | 1.35 | 2.06 | 0.41 | 0.19 | -1.07 | |
| Leucine-rich repeat/extensin | 0.74 | 3.40 | 2.20 | 1.67 | 0.78 | -1.10 | |
| Purine transporter | 0.27 | 1.22 | 2.19 | 3.90 | 1.70 | -1.20 | |
| B3 domain-containing protein | 0.11 | 0.52 | 2.31 | 0.47 | 0.37 | -0.34 | |
| Late blight resistance protein | 0.95 | 2.22 | 1.22 | 1.37 | 0.77 | -0.84 | |
| Disease resistance protein | 15.43 | 5.83 | -1.40 | 3.14 | 13.77 | 2.13 | |
| NBS-LRR type disease resistance protein | 1.73 | 0.51 | -1.76 | 1.27 | 1.59 | 0.32 | |
| Pyruvate decarboxylase | 494.52 | 80.00 | -2.63 | 12.71 | 88.10 | 2.79 | |
| Phospholipase A1 | 1.65 | 12.81 | 2.96 | 0.56 | 3.88 | 2.79 | |
| LRR receptor-like serine/threonine-protein kinase | 7.41 | 44.71 | 2.59 | 2.09 | 8.29 | 1.99 | |
| Leucine-rich repeat transmembrane protein kinase | 11.27 | 56.82 | 2.33 | 2.39 | 11.21 | 2.23 | |
| Protein kinase | 4.54 | 24.39 | 2.42 | 0.68 | 3.79 | 2.48 | |
| Germin 3 | 270.46 | 1189.43 | 2.14 | 8.24 | 127.97 | 3.96 | |
At the cell wall level, plants develop modifications to prevent the entry of biotic pathogen agent. One such case is expression of pectinasterase (locus ST4.03ch03: 39485109-39486959) which was higher in “María Huanca” (Table 3). Validating the research by Uehara et al. (2007), who found a tomato (Solanum lycopersicum) variety resistant to Globodera rostochiensis. This variety showed high levels pectinasterase expression. Germin and Lemir genes reached a higher value of RPKM after infection of Globodera pallida, compared to the susceptible variety. Germin (locus ST4.03ch01: 78647674-79648034) is an enzyme with oxalate oxidase activity, producing hydrogen peroxide (H2O2), toxic to pathogenic agents (Lane et al. 1994, Caliskan 2000, Walter, et al. 2018). On the other hand, Lemir has an anti- insect/anti pathogenic action, implicated in with defense against nematodes or other pathogens (Ryan 1990).
Blight resistance protein Rpi-blb2, TMV resistance protein N, Tospovirus resistance protein C, RPI protein, late blight resistance protein, Cc-nbs-lrr resistance protein, R3a disease resistance protein, Nbs-lrr resistance protein, and HJTR2GH1 protein were found expressed in the root transcriptome of Solanum phureja confronted with Globodera rostochiensis,Kochetov (2017). In wild potato relatives resistance genes were identified as Gro1-4, which confer resistance to all the pathotypes of G. rostochiensis (Paal et al. 2004). These are part of the NBS-LRR family and the Grp1 gene for G. pallida (Finkers-Tomczak et al. 2009)
Metabolic pathways related to defense against Globodera pallida were found in “Maria Huanca”. Protein kinases (Table 3) are a central component in phosphorylation cascades of extracellular signal transduction proteins. This activity could be related to an expression of certain R genes such as: NBS-LRR type disease resistance protein, Late blight resistance protein, TMV resistance protein, Nematode resistance, Disease resistance protein, all of which were expressed with greater predominance in “Maria Huanca”. In the case of these R genes, expression levels were very low, corroborating the assertions by Hammond- Kosack and Jones (1997) and Hulbert et al. (2001) who detailed that R genes are expressed constitutively at very low levels, even before the appearance of the pathogen or during infection. This implies that R genes are being expressed themselves constitutively; giving plants a system of permanent alert to potentially invasive pathogens. These expression levels of R genes may be sufficient to mediate resistance to the invading pathogen (Lozano et al., 2015). Globally, results demonstrate differential expression of genes related to the infection response of G. pallida, between resistant and susceptible potato variety. The function of these genes corroborates their participation in defense against Globodera pallida, and therefore their relationship with the resistance response.
On the other hand, some of these genes were also overexpressed in the susceptible variety, but not sufficient quantities as to confer absolute resistance. It is conceivable that there are genes such as microRNAs that are regulating the expression of these genes, or post- transcriptional changes that condition the variation in the expression of each gene identified as related to the resistance against G. pallida.











 uBio
uBio