Introduction
Peru is one of the most biodiverse countries in the world. Current estimates of species richness showed the occurrence of high numbers of several taxa, such as plants (20533), vertebrates (5738), and arthropods (30547) (MINAM, 2019). Conservation and management of this vast biodiversity requires different approaches considering ecological and systematic knowledge, and, recently, information about genetic diversity (Noreña et al. 2018). This information is essential not only for conservation and taxonomic studies but also for bioprospecting novel compounds (Sekurova et al. 2019) or for breeding programs assisted by genetic markers (Assefa et al. 2019, Mrode 2019).
Bioinformatics, the discipline that uses computational approaches to answer biological questions, has become essential to analyze genetic and genomic information (Baxevanis & Ouellette 2005), and to study large DNA datasets obtained via next-generation sequencing (NGS) to discover new genes, functions, pathways, and molecular interactions. Most of this information is available in public online databases. For instance, GenBank (https:// www.ncbi.nlm.nih.gov/genbank), at the National Center for Biotechnology Information from the US National Institute of Health, currently holds 216531829 sequences, being one of the largest sources worldwide. Other databases such as the Barcode of Life Data System (https:// www.boldsystems.org), are more specific, providing information from specific eukaryotic genes.
The representativeness of Latin American data in genome projects and public databases has been discussed since the beginning of the genomic era (Ramírez et al. 2002). The information stored in databases has become essential to study the genetic diversity of indigenous peoples (Harris et al. 2018), human diseases (Norris et al. 2018), and regional biotechnological and agro-industrial challenges (Sasson & Malpica 2018, Wang et al. 2017).
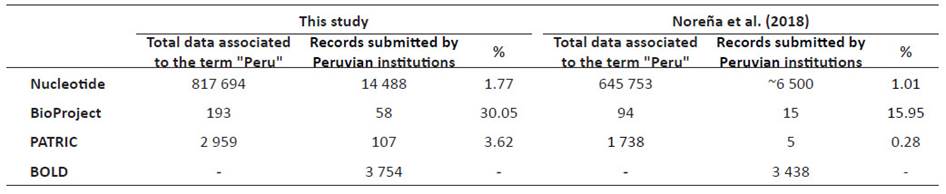
The most recent assessment of the Peruvian genetic data in public databases was done by Noreña et al. (2018). The authors estimated 645753 sequences in the NCBI Nucleotide database associated with the term "Peru", from which 6522 (1.01%) were published by national institutions. We updated the amount of sequencing data of the Peruvian biodiversity submitted by national institutions to four main public genetic databases: NCBI BioProject, a database that comprises multiple collections of biological data related to a single genomic sequencing initiative, NCBI Nucleotide that comprises GenBank and related databases, the exclusively bacterial database Pathosystems Resource Integration Center (PATRIC) which provides sequencing, protein-protein interactions, and transcriptomic data (Wattam et al. 2017), and the Barcode of Life Data System (Ratnasingham & Hebert, 2007), a DNA barcode database, focused on single markers, namely, the cytochrome oxidase I (COI) gene for animals, internal transcribed spacers (ITS) for fungi, and the ribulose-bisphosphate carboxylase (rbcL) and maturase K (matK) for plants. The same databases were used by Noreña et al. (2018).
Material and methods
Nucleotide data was retrieved using Entrez Direct (Kans 2013). In April 2020, we downloaded all the records from the Nucleotide database containing the query "Peru" using the following command,
esearch -db nucleotide -query "Peru" | efetch -format gb > peru.gb
The output of this search, a 30 Gb file in Genbank format, was used to extract information such as the journal and organism of the published sequence. In Nucleotide, information of the institution that submitted the genetic sequence is stored in the "journal" variable. A file containing only unique journal names was also created to count all Peruvian institutions in the database. Through awk scripts, we counted how many times the word "Peru" appeared in the variable journal, how many times a certain institution uploaded sequences to this database, and how many organisms have been sequenced. We also analysed which organisms were the most sequenced by each institution in Peru.
BioProject data was also retrieved using Entrez Direct and the command,
esearch -db BioProject -query "Peru" | efetch -format xml | xtract -pattern DocumentSummary -element Project \ -block Organism -element OrganismName Supergroup \ -block Submission -element Name > BioProject.xml
and processed using awk scripts. Both, BOLD and PATRIC data were retrieved directly from their webpages, using "Peru" as a search query. This data was downloaded in tabular (tsv) and comma-separated (csv) formats, respectively. For both datasets, cat and grep scripts were used. cat concatenates texts to create a new output, and grep -c, counts the occurrences of a certain word query. awk scripts were additionally used for the BOLD dataset to group and count most sequenced taxonomic orders per institution. For the PATRIC dataset, csvgrep was used to count sequenced organisms per institution. All code and scripts are available in this GitHub repository: https://github.com/reymonera/mining_peru_sequence_DB. Finally, we searched in the Scopus reference database (https://www.scopus.com) for records with the term "genetic diversity" produced by at least one Peruvian institution.
Results
Our results are summarized in Figure 1 and Table 1. In addition, complete information of the records from each database described here can be found in the Supplementary Information associated with this publication (Romero & Castillo-Vilcahuaman, 2020). We found 817694 records associated with the term "Peru'' in the Nucleotide database (Table 1). However, this number could be an overestimate of Peruvian sequences in this database. For instance, we found some records that corresponded to the Spanish institution Estación Biológica de Doñana, because the building where it is located is named "Pabellón del Peru". Thus, to be consistent with the previous report from Noreña et al. (2018), we only focused on submissions by Peruvian institutions. Therefore, we found 14 488 sequences (1.77% of the records related to the term "Peru") submitted by 36 Peruvian institutions (Suppl. Inf. 2). 11 institutions submitted more than 100 records to this database (Fig. 1). Public institutions such as Instituto Nacional de Salud (INS), Universidad Nacional Mayor de San Marcos (UNMSM), Instituto Nacional de Investigación Agraria (INIA), and Instituto de Investigaciones de la Amazonía Peruana (IIAP), and private institutions such as Universidad Peruana Cayetano Heredia (UPCH) and Farmacéuticos Veterinarios S. A. C. (FARVET) submitted most of the records. In BioProject, we found 193 records related to the term "Peru" (Suppl. Inf. 3), 59 of them were submitted by 9 Peruvian institutions (Fig. 1). INS has most of the records, followed by UNMSM, Universidad Nacional Agraria La Molina (UNALM) and UPCH. In PATRIC, we found 2959 records (Suppl. Inf. 4), 107 of them submitted by 8 Peruvian institutions (Fig. 1). Similarly, most of the records were submitted by UPCH, INS, and UNMSM. Finally, in BOLD, we found 23968 public records associated to the term "Peru".
From these, 8249 were mined from Genbank so to avoid counting them twice, we discarded them in the final count. We found that 3754 records were submitted by 11 Peruvian institutions (Suppl. Inf. 5). Most of the sequences were produced by the Centro de Ornitología y Biodiversidad (CORBIDI), UNMSM (mainly, from the Museo de Historia Natural), UNALM (mainly, from the Herbarium), the Instituto del Mar del Perú (IMARPE), and the Servicio Nacional de Sanidad Agraria del Perú (SENASA).
Records from the Nucleotide, BioProject and PATRIC databases are biased to pathogens probably reflecting the research field of groups that frequently submit DNA data and the direction of research funding. In the Nucleotide database, most records belong to Pasteurella multocida, and Neisseria meningitidis, followed by Vibrio parahaemolyticus, Bacillus thuringiensis, Escherichia coli, Shewanella sp., Aeromonas veronii, Homo sapiens, HIV, and Mycobacterium tuberculosis (Figure 2). 70.60% of the records belonged to Bacteria. 24.38%, to Eukaryota and 3.07%, to Viruses (Suppl. Inf. 2). In addition, we found no records of Archaea submitted by Peruvian institutions. Within Eukaryota, most of the records belonged to clades Fungi and Viridiplantae and Phyla Chordata, Arthropoda, and Mollusca (Suppl. Inf. 2). In BioProject, Salmonella enterica subsp. enterica serovar Infantis is the most frequent record (Suppl. Inf. 3). PATRIC’s most frequent records are pathogenic bacteria, namely, Mycobacterium tuberculosis, Yersinia pestis, Shigella sonnei, and Staphylococcus aureus (Suppl. Inf. 4). BOLD presents different results because it is a Eukaryotic database. BOLD’s most frequent taxa are, in descending order, Chordata (Aves and Actinopterygii), Angiospermae, and Arthropoda (Insecta, and Arachnida) (Suppl. Inf. 5).
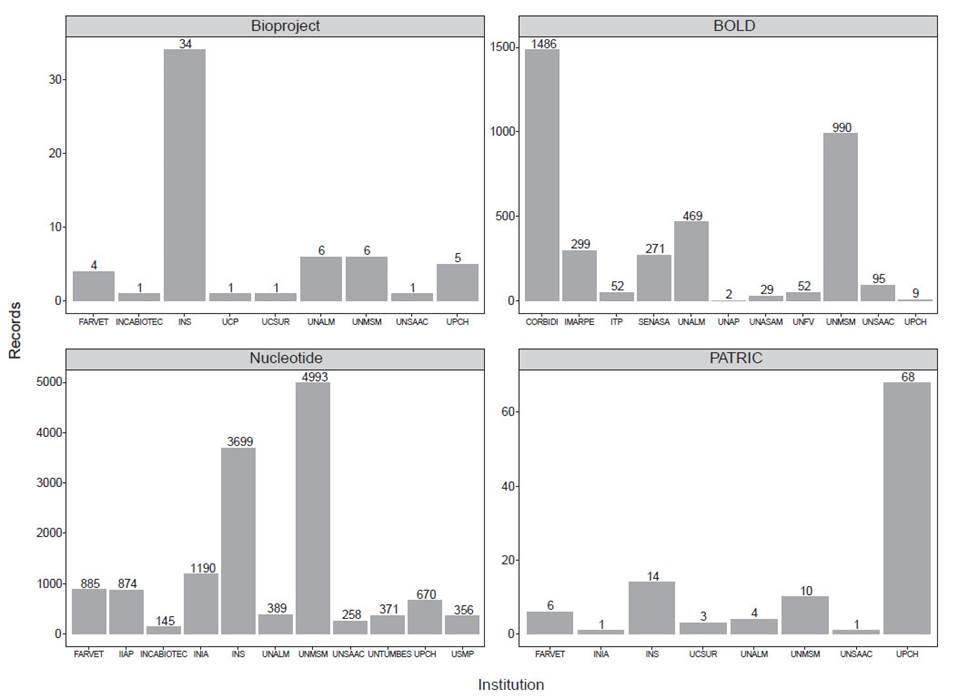
Figure 1 Peruvian institutions that submitted most of the records to four public genetic databases; BioProject, BOLD, Nucleotide and PATRIC. For Nucleotide, we show institutions that submitted more than 100 records to this database.
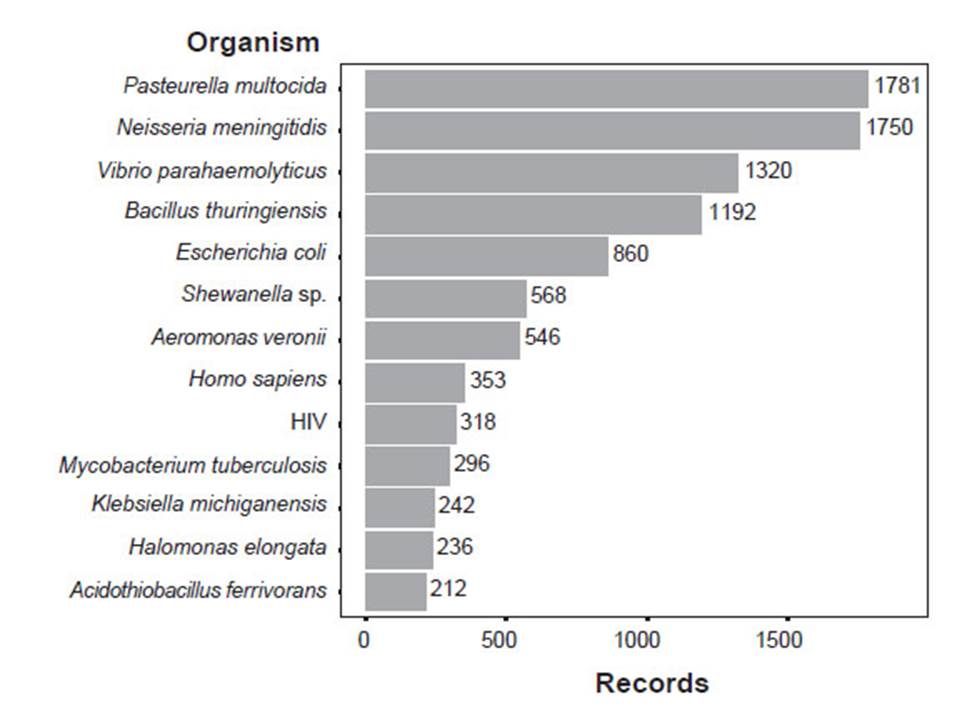
Figure 2 Most abundant records of organisms submitted by Peruvian institutions to the Nucleotide database. The figure shows organisms with more than 200 records.
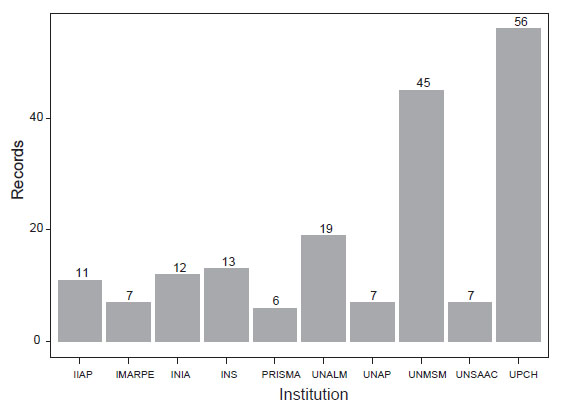
Figure 3 Peruvian institutions that frequently appeared in scientific articles about genetic diversity. The literature and affiliation search were done in Scopus
We found 266 articles published from authors affiliated to at least one Peruvian institution (Figure 3, Suppl. Inf. 6). Results were similar to the previous ones: UPCH, UNMSM, UNALM, and INS appeared in most of the results. Research groups collaborated mainly with institutions from the United States, Brazil, United Kingdom, Colombia, and France. Main subject areas in these investigations are Agricultural and Biological Sciences, (Bio) Medicine and Environmental Sciences (Suppl. Inf. 7)
Discussion
It has been more than two years since Noreña et al. (2018) reported the first results of Latin American genetic data stored in public databases. These authors kindly shared their scripts and information with us to reproduce and compare results. In our view, open science and data must be promoted, thus, all our scripts and results can be freely accessed and downloaded from GitHub.
Noreña et al. (2018) reported 1.01% (~6500) of records deposited by Peruvian institutions in NCBI Nucleotide. We showed an increase of more than two times (14488) in records in only two years (Table 1). Although, our calculated occurrence (1.71%) did not change a lot because the permanent increase of Nucleotide total records. An independent report by Clark et al. (2016) compared GenBank information between a similar range of time (20142016) and found two to four-fold increases in both invertebrate and vertebrate data. In the case of Peru, INS and UNMSM have generated more data than all other institutions together (> 8000 records). In BioProject we found 30.05% records submitted by Peruvian institutions (58 out of 193) doubling the occurrences found by Noreña et al. (15.95%). In PATRIC, Noreña et al. (2018) found 0.28% records submitted by Peruvian institutions while we found 3.62% records (107 out of 2959). Finally, in BOLD, we reported 3754 records while Noreña et al. (2018) found 3438. In the latter case, the increase was smaller than in the other databases.
It is not a surprise that the same institutions are actively submitting data to different databases because they host strong research groups related to emerging and infectious diseases, biodiversity, and biotechnology. However, most of them are based in Lima, capital of Peru. We believe that, to increase diversity and representativeness, funding efforts should be directed to more regional research groups and other highly diverse taxa, for instance, non-pathogenic bacteria, archaea, non-vertebrates, and non-flowering plants.
We are aware of the limitations of our searches. We limited the search to the four databases mentioned above for consistency and reproducibility. We also limited the search to nucleotide sequences. Further information of protein sequences and structures can be found in the databases NCBI Protein and Protein Data Bank (Berman et al. 2000). Also, as we consider only Peruvian institutions, we did not mention international institutions based in Peru, for example, the Centro Internacional de la Papa (CIP) or the US Naval Medical Research Unit Six (NAMRU-6) that also produced sequences from Peruvian organisms.
However, as both institutions are based in Lima, their inclusion did not change the main conclusions of our analysis (see Suppl. Inf. 2, 3, 4, 5). Furthermore, we relied on the information provided by the researchers which could result in heterogeneous data even if it comes from the same institution, for instance, in BioProject, we found records from INS, with different names such as "Peruvian National Institute of Health", "Instituto Nacional de Salud-Peru", "Instituto Nacional de Salud" (Suppl. Inf. 3). Thus, we believe that each institution must develop a homogenous protocol for data submission.
Sequencing projects will definitely rise in the next months and years. It would not be a surprise, for instance, to see a rise of SARS-CoV-2 genomes submitted by Peruvian institutions. In order to effectively track this information and keep it updated we encourage Peruvian institutions to create a unified online platform to frequently survey and organize sequencing data.
Table 1 Number of records associated to the term "Peru" and submitted by Peruvian institutions in four genetic public databases.
We agree with several members of the scientific community that in order to improve the quality of science: transparency, reproducibility, efficiency, and benefits to society, there is a need for open data (Molloy 2013). Public databases play a big role in open science and reproducibility. Thus, the availability of sequencing data in this platform is crucial for scientific research and should be practiced and encouraged. In addition, we believe in the necessity of a national genome sequencing plan developed by public and private research institutions.
This will increase the Peruvian genetic information in public databases, articulate efforts and promote networking from different institutions, and provide resources to manage issues from several topics such as agriculture, public health and biodiversity conservation. Finally, efforts should be also focused on a country-wide capacity building initiative to train a new generation of Peruvian bioinformaticians that will be essential in future challenges to our biodiversity











 uBio
uBio