Serviços Personalizados
Journal
Artigo
Indicadores
-
 Citado por SciELO
Citado por SciELO
Links relacionados
-
 Similares em
SciELO
Similares em
SciELO  uBio
uBio
Compartilhar
Revista Peruana de Biología
versão On-line ISSN 1727-9933
Rev. peru biol. vol.20 no.2 Lima ago. 2013
TRABAJOS ORIGINALES
Detection of Bartonella spp. and Rickettsia spp. in fleas, ticks and lice collected in rural areas of Peru
La detección de Bartonella spp. y Rickettsia spp. en pulgas, garrapatas y piojos recolectados en las zonas rurales de Perú
Abraham G. Cáceres1, 2, Carlos P. Padilla Rojas3, Javier Arias Stella4, Gerardo Huatuco Crisanto5, Antero Gonzales Pérez6
1 Departamento de Microbiología Médica, Facultad de Medicina Humana, Universidad Nacional Mayor de San Marcos, Lima - Perú. E-mail Abraham G. Cáceres: acaceres31@hotmail.com
2 Laboratorio de Entomología, Instituto Nacional de Salud, Lima - Perú
3 Laboratorio de Biotecnología y Biología Molecular, Instituto Nacional de Salud, Lima - Perú
4 Instituto de Patología y Biología Molecular Arias Stella, Lima - Perú
5 Centro de Salud de San Ignacio, Sub Región de Salud Jaén, Dirección Regional de Salud Cajamarca, Caja-marca - Perú
6 Hospital de Apoyo Santiago Apóstol Utcubamba, Red de Salud Unidad Ejecutora 404 Utcubamba, Dirección Regional de Salud Amazonas, Amazonas - Perú
Abstract
Bartonellosis and rickettsiosis are commonly reported in Peru. In order to detect Bartonella sp. and Rickettsia sp. in fleas, ticks and lice, specimens from five distinct locations in Peru (Marizagua, Cajaruro, Jamalca, Lonya Grande and El Milagro) were collected and screened for the presence of these bacteria using PCR and later confirmation by DNA sequencing. The specimens collected were distributed in 102 pools (76 Ctenocephalides felis, 2 Ctenocephalides canis, 16Pulex irritans, 5 Pediculus humanus, 2 Rhiphicephalus sanguineus, and 1 Boophilus spp.), whereBartonella was detected in 17 pools (6 of C. felis, 9 of P. irritans, 1 of C. canis, and 1 P. humanus). Also, Rickettsia was detected in 76 pools (62 C. felis, 10 P. irritans, 2 P. humanus, and 2 C. canis). Bartonella clarridgeiae was detected in C. felis, C. canis and P. irritans pools at 5.3%, 50% and 12.5%, respectively. Bartonella rochalimae was detected in one C. felis and two P. irritans pools at 1.3% and 12.5%, respectively. Furthermore, B. henselae was detected in one C. felis pool and one P. humanus pool corresponding to 1.3% and 20%, respectively; andBartonella spp. was also found in 5 pools of P. irritans at 31.3%. Additionally, R. felis was detected in C. felis, C. canis and P. irritans pools at 76.3%, 100% and 37.5%, respectively; and Rickettsia spp. was detected in C. felis, P. irritans and P. humanus pools at 5.3%, 25% and 40%, respectively. These results demonstrate the circulation of these bacteria in Peru.
Keywords: Bartonella ; Rickettsia; arthropods; PCR detection; Peru.
Resumen
La Bartonellosis y la Rickettsiosis son enfermedades comúnmente reportadas en Perú. Con el propósito de detectar Bartonella sp. y Rickettsia sp. especímenes de pulgas, garrapatas y piojos de cinco localidades del Perú (Marizagua, Cajaruro, Jamalca, Lonya Grande and El Milagro) fueron colectadas y analizadas. Para la detección se usó PCR y una posterior confirmación con secuenciamiento de DNA. Los especímenes colectados fueron agrupados en 102 pools (76 Ctenocephalides felis, dos Ctenocephalides canis, 16Pulex irritans, cinco Pediculus humanus, dos Rhiphicephalus sanguineus, y un Boophilus spp.). Bartonella fue detectada en 17 pools (seis de C. felis, nueve de P. irritans, uno de C. canis, y uno de P. humanus). Rickettsia fue detectada en 76 pools (62 de C. felis, 10 de P. irritans, dos de P. humanus, y dos de C. canis). Bartonella clarridgeiae fue detectada en C. felis (5.3% especímenes), C. canis (50%) y P. irritans (12.5%). Bartonella rochalimae fue detectada en C. felis (1.3%) y P. irritans (12.5%). Además, se detectó B. henselae en C. felis (1.3%) y P. humanus (20%). Bartonella spp. también se encontró en P. irritans (31,3%). Además, se detectó R. felis en C. felis (76.3%), C. canis (100%) y P. irritans (37.5%), y Rickettsia spp. se detectó en C. felis (5,3%), P. irritans (25%) y P. humanus (40%). Estos resultados demuestran la circulación de estas bacterias en el Perú.
Keywords: Bartonella ; Rickettsia; arthropods; PCR detección; Perú.
Introduction
Human bartonellosis is caused by various species of Bartonella. Nowadays, there are several species of this genus that can infect humans including B. bacilliformis, B. henselae, B. quintana, B. rochalimae, B. elizabethae, B. vinsonii subsp. arupensis, B. vinsonii subsp. berkhoffii, B. grahamii, B. koehlerae, B. washoensis, B. alsatica, and B. tamiae (Billeter et al. 2008, Daly et al. 1993, Kerkhoff et al. 1999, Roux et al. 2000, Eremeeva et al. 2007, Kosoy et al. 2008). Only B. bacilliformis, B. vinsonii, B. quintana, B. rochalimae, B. elizabethae and B. cladrrigeiae have been reported in Peru (Billeter et al. 2008, Eremeeva et al. 2007, Parola et al. 2002, Roux & Raoult 1999, Raoult et al. 1999). Bartonella is believed to be transmitted by an arthropod vector, and infection can result in Carrions disease, cat scratch disease, trench fever, endocarditis, neuroretinitis, bacillar angiomatosis, hepatic granulomatosis, arthritis, osteomelitis, and sepsis (Billeter et al. 2008, Breitschwerdt & Kordick 2000).
Rickettsia spp. are gram negative coco-bacilli and intracellular pathogens and most of them are transmitted transtadially by a wide variety of arthropod hosts including fleas, lice and ticks. Infections with these pathogens can cause diverse human diseases such as endemic typhus, scrub typhus, erhlichiosis and others rickettsiosis (Raoult & Roux 1997). In Peru, rickettsiosis occurs sporadically along the coast, highlands and jungle. The exposure to rickettsiosis was confirmed by immunofluorescence assay (IFA) in regions of Peru (Raoult et al. 1999, Blair et al. 2004a) and Rickettsia species were detected in ticks and fleas (Schoeler et al. 2005, Jiang et al. 2005).
Several arthropods belonging to the orders Diptera (Noguchi et al. 1929, Caceres et al., 1997), Anoplura (Ellis et al. 1999), Siphonaptera (Brouqui et al. 1999; Higgins et al. 1996) and Parasitiformes (Schouls et al. 1999, Angelakis et al. 2010) have been implicated in the transmission of Bartonella spp. Previous experimental studies have demonstrated that Lutzomyia verrucarum (sandfly) are competent vectors of B. bacilliformis, which causes Carrions disease (Breitschwerdt & Kordick 2000, Maguiña et al. 2009). In order to detect Bartonella spp. and Rickettsia spp. in fleas, lice and ticks, specimens were captured from humans, domestic and wild animals in five distinct locations in Peru, where Carrions disease is endemic. The presence of Bartonella spp. in these specimens was evaluated by polymerase chain reaction (PCR) and confirmed by DNA sequencing.
Materials and methods
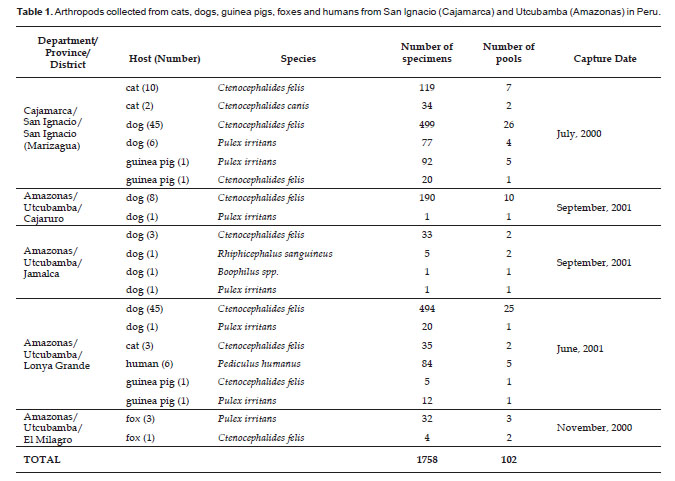
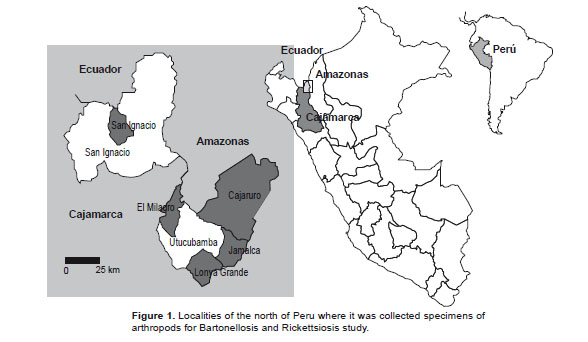
Study sites and specimen collections. Samples were collected from random houses in different places: San Ignacio (Marizagua) of Cajamarca departament; Cajaruro, Jamalca, Lonya Grande and El Milagro of Amazonas departament in July 2000 to September 2001 (Table 1, Fig. 1).
These samples were obtained from different pets such as dogs, cats, and guinea pigs (considered as a food source); moreover, foxes found near the neighborhood were trapped in cages for later sample collection. Specimens such as fleas were collected by hand from animals; but lice were collected from people (head, body and clothes) and ticks were collected only from dogs. These were then stored in vials containing 70% ethanol, and organized according to host and place of collection.
The taxonomic classification was done in the Entomology Laboratory at the Daniel A. Carrión Tropical Medicine Institute, of the Universidad Nacional Mayor de San Marcos in Lima, using entomological keys (Jonson 1957, Lewis 1972).
After taxonomic classification, the specimens were grouped into pools of 6 to 20 specimens, each pool from one or two hosts, to reduce the processes for analysis (see Table 1).
DNA extractions from specimens. The whole specimens were grouped in pools and washed with 1X Phosphate Buffered Saline (PBS). They were then triturated with a micro-pestle in 100 µL of PBS. DNA was extracted using a Tissue DNA Purification Kit (QIAGEN) following the standard procedure recommended by the manufacturer.
DNA amplification of the gltA gene by PCR. The gltA gene of Bartonella was amplified using the BhCS.781p and BhCS.1137 primers (Norman et al. 1995). The same specimens were analyzed using the Rp877p and Rp1258n primers, theses primers amplified the gltA gene of Ricketsia (Regnery et al. 1991).
The amplification reactions consisted of 5 µL of amplification buffer 10X, 5 µL (10 µM) of each primer, 5 µL (25 mM) of magnesium chloride, 2 µL (10 mM) of nucleotide triphosphates, 0.2 µL (5 UµL) of AmpliTaq Gold DNA polymerase (Applied Biosystems Inc.) and 5 µL of purified DNA to each tube. The final volume of the amplification reaction was 50 µL. Purified DNA from B. bacilliformis strain ATCC 35685 and a Rickettsia felis characterized culture were used as positive controls. The amplifications were performed on a 9700 Applied Biosystems thermocycler, and the cycles were as follows: 1 cycle for 95 °C for 10 min; 35 cycles at 95 °C for 0.5 min, 50 °C for 0.5 min, and 72 °C for 1 min; 1 cycle at 72 °C for 5 min; and a final soaking at 4 °C until analysis.
The amplified products were analyzed by electrophoresis in agarose gels, stained with ethidium bromide and visualized in a UV transilluminator.
The amplified products were then purified using QIAQuick Gel extraction Kit (QIAGEN Inc.) and sequenced using a Big Dye DNA terminator Kit (Applied Biosystems Inc.) on an ABI310 sequencer (Applied Biosystems) at the Peruvian National Institute of Health and following standard procedures. The sequences obtained were analyzed with GenBank using BLAST software (www.ncbi.nlm.nih.gov/BLAST). Clustalw software (http://align.genome.jp/) was used for alignment and comparison of sequences.
Results
A total of 1758 specimens were captured from humans, cats, dogs, guinea pigs and foxes (Table 1). Of these, 1399 (79.58 %) samples were identified as Ctenocephalides felis (collected from dogs, cats, guinea pigs and foxes). 235 (13.37%) specimens were Pulex irritans (collected from dogs, guinea pigs and foxes); 84 specimens (4.78%) were Pediculus humanus (collected from humans); 34 specimens (1.93%) were Ctenocephalides canis (collected from cats); 5 specimens (0.28%) were Rhiphicephalus sanguineus (collected from dogs); and 1 specimen (0.06%) was identified as Boophilus spp. (collected from a dog) (Table 1).
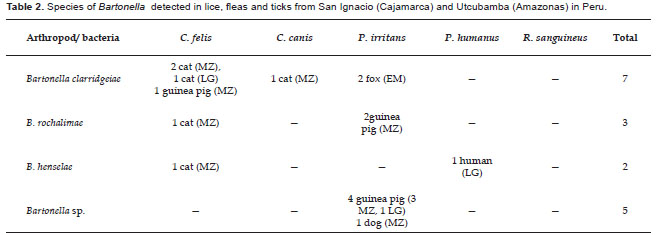
One hundred two pools were processed by PCR. Of these, 17 pools (16.67%) were PCR-positive using primers for Bartonella genera. These pools were confirmed as Bartonella by DNA sequencing.
Bartonella clarridgeiae (accession number AY836149) was detected in 4 pools of C. felis collected from three cats and one guinea pig, 1 pool of C. canis collected from a cat and 2 pools of P. irritans collected from a fox (all 100% homologous with each other). The AY836149 nucleotide sequence was compared with GenBank Database showing 100% homology with B. clarridgeiae Houston-2 cat strain (U84386) (Table 2).
Bartonella rochalimae was detected in 1 pool of C. felis collected from a cat and 2 pools of P. irritans collected from guinea pigs (all 100% homologous to GenBank accession number GU583842). The GU583842 nucleotide sequence was 100% Bartonella henselae was detected in 1 pool of C. felis collected from a cat and 1 of P. humanus collected from humans (accession numbers AY840994 and GU583844). Both AY840994 and GU583844 nucleotide sequences showed 100% homology with B. henselae strains: BR01 (GU056191), BCF01 (GU056188) and CS3 (FJ832097) (Table 2).
Bartonella spp. was detected in 5 pools of P. irritans: four pools collected from guinea pig and one from a dog, all 100% homologous to GenBank accession N° GU583846. The GU583846 nucleotide sequence showed 96.3% and 96.6% homology with Bartonella spp. DNA detected in a flea and blood collected from Sigmodon hispidus with GenBank accession numbers EF616666 and AF082322, respectively (Table 2).
Seventy-six pools (74.51%) were PCR positive using primers for Rickettsia. Of these, 66 pools were positive for Rickettsia felis (58 pools of C. felis, 2 pools of C. canis and 6 pools of P. irritans; all 100% homologous to GenBank no. GU583847). The GU583847 nucleotide sequence showed 100% homology with Rickettsia felis collected from C. felis (EU853837), Liposcelis bostrychophila (GQ329877, GQ329873), and Pulex echidnophagoides (GU447233) (Table 3).
Ten pools were positive for Rickettsia spp. (4 pools of C. felis, 4 pools of P. irritans and 2 pools of P. humanus), where one of the pools was reported as GenBank no. GU583848 and the other nine pools were 100% homologous to GU583849 (Table 3). The nucleotide sequence GU583848 showed 100% homology with Rickettsia spp. collected from Pulex irritans (EU853838), Echidnophaga gallinacea (DQ166938), C. canis (AF516333) and C. felis (AY953289); also the nucleotide sequence GU583849 showed 100% homology with Rickettsia sp. collected from C. felis (AY953288) and C. canis (AF516331). DNA of Bartonella and Rickettsia werent detected in ticks.