Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO
Related links
-
 Similars in
SciELO
Similars in
SciELO
Share
Revista de Gastroenterología del Perú
Print version ISSN 1022-5129
Rev. gastroenterol. Perú vol.33 no.2 Lima Apr./Jun. 2013
ARTÍCULO DE REVISIÓN
Targeting the microbiota in the management of gastrointestinal and liver disease
Apuntando a la microbiota en el manejo de las enfermedades gastrointestinales y hepáticas
Eamonn MM Quigley 1ab, Howard P Monsour 1a
1 Gastroenterology and Hepatology, The Methodist Hospital and Weill Cornell Medical College
a MD; b FRCP FACP FACG FRCPI
ABSTRACT
Thanks to rapid advances in technology the details of the human microbiome and its functions in health and disease are being progressively revealed. Though many reports have linked various disease states with an altered microbiome and while some associations between the microbiome and disease states are well established, many of these studies are largely descriptive and the changes reported in the microbiome have yet to be shown to be causative. A number of strategies are available to modify the microbiota; some such as the use of antibiotics for specific indications, are well established, others such as the use of probiotics and prebiotics in a variety of disease states are supported by more limited data. Fecal transplantation has emerged as an exciting, albeit rather drastic, intervention for intestinal and, perhaps, other disorders. Other approaches, such as the isolation, purification and formulation of small molecules with specific biological actions, derived from the microbiota look very promising.
Key words: Gastrointestinal diseases; Intestinal diseases; Liver diseases (source: MeSH NLM).
RESUMEN
Gracias al rápido avance de la tecnología los detalles del microbioma humano y sus funciones en salud y enfermedad están siendo conocidos progresivamente. A pesar que muchos reportes han relacionado varios estados de enfermedad con un microbioma alterado y mientras algunas asociaciones entre el microbioma y estados de enfermedad están bien establecidas, muchos de estos estudios son solo descriptivos y los cambios reportados en el microbioma todavía tienen que demostrarse que son la causa. Existen muchas estrategias para cambiar la microbiota; algunas como el uso de antibióticos para indicaciones específicas, están muy bien determinadas, otras, como el uso de probióticos y prebióticos en una gran variedad de enfermedades, están sustentadas en data más limitada. El trasplante fecal ha surgido como una alternativa muy emocionante, aunque algo drástica, para las enfermedades intestinales y quizás también para otras patologías. Otros abordajes como el aislamiento, purificación y formulación de pequeñas moléculas con acciones biológicas específicas, derivados de la microbiota aparecen como muy prometedoras.
Palabras clave: Enfermedades gastrointestinales; Enfermedades intestinales; Enfermedades del hígado (fuente: DeCS BIREME).
BACKGROUND
Due largely to rapidly evolving advances in analytical techniques in microbiology, molecular biology and bioinformatics (1-3) the true diversity of the population of micro-organisms that inhabits the gastrointestinal tract of man (collectively referred to as the human gut microbiota) is being revealed and its contributions to homeostasis in health and to the pathogenesis of disease appreciated. Formerly, bacteria in the gastrointestinal tract had to be individually identified by tedious and cumbersome culture-based techniques and, while it is theoretically possible to characterize the entire population of bacteria in the gut employing this approach, the resources and time involved would be absolutely prohibitive. The advent of high-throughput techniques based on molecular methodologies has revolutionized the field and now permits the rapid identification of all species and strains from a given sample (1-3); these methodological advances (coupled with dedicated bioinformatics expertise) are largely responsible for the study of the microbiota being currently one of the most active and exciting in all of science. One has only to glance through the pages of such major scientific journals as Nature and Science to appreciate the prominence of this field. Major collaborative projects have been initiated to assess the variability of the microbiota across various populations in health (4-7) and relationships to phenotype in diseases as varied as diabetes (8,9) and cystic fibrosis (10).
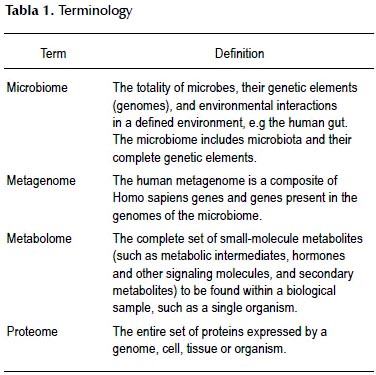
The application of molecular techniques to the study of microorganisms has generated a whole new terminology; the major terms are listed and defined in Table 1. It should be stressed that the term microbiome is now to be preferred to the older term "flora" as, unlike the latter, the microbiome includes viruses, fungi and other microorganisms which are clearly normal residents of the human gut. It is estimated that the intestinal microbiota composed of 1013 to 1014 microorganisms; whose microbiome contains more 100 times as many genes as the human genome. Given this discrepancy one could reasonably ask: who is the host? While the emphasis in this review will be on the composition of the gut microbiota and how it may be modified, it should be noted that the study of the metabolome (metabolomics) is a rapidly advancing field which is revealing what the microbiome actually does.
The microbiota in health
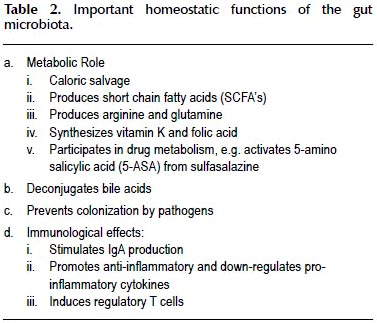
The contributions of the microbiota ("the forgotten organ") to homeostasis in health are being more completely understood and appreciated. Some of these functions, such as the synthesis of vitamin K and folate, deconjugation of bile acids and production of short chain fatty acids are well known (11); others are more recent revelations (12). Some of the main functions of the microbiota are listed on Table 2.
Central to the beneficial interaction between the microbiota and man is the manner in which bacteria and, most likely, other microorganisms, contained within the gut "talk" to the hosts immune system and participate in a variety of metabolic processes of mutual benefit to the host and the microbe. Indeed, through the development of tolerance, the microbiota and the host learn to live together in a symbiotic relationship (13,14). These immunological effects are mediated, at least in part, via dendritic (antigenpresenting cells, APCs) cells (15). Interactions with the mucus layer and the epithelial barrier contribute to the maintenance of intestinal barrier integrity.
More recently the metabolic functions of the microbiota have received considerable attention in large part related to observations linking alterations in the microbiota to obesity and such complications as diabetes and the metabolic syndrome. The essential observations here were, firstly, that the gut microbiota of the genetically obese ob/ob mouse contained 50% fewer Bacteroidetes and more Firmicutes than a lean mouse (16), secondly, than an obesity-inducing diet decreased the diversity of the microbiota by decreasing the proportion of Bacteroidetes and increasing that of Mollicutes (17) and, thirdly and most critically, that an obese microbiome was highly efficient at extracting energy (18,19). While the contribution of the microbiome to obesity in animal models and in man continues to be explored (20,21), others have focussed on the ability of components of the microbiome to induce subtle and highly specific alterations in the composition of various organs and tissue compartments (22-26). The demonstration that the oral administration of a single bacterial strain with commensal properties can induce compositional changes in organs as remote as the brain is a fascinating illustration of the potent effects of the microbiota (25,26). The idea that the microbiome could interact with distal organs has been taken a step further in the concept of the microbiome-gut-brain axis. This is based on a number of observations in a number of animal models which have shown how manipulation of the microbiota can alter behaviour, brain function and morphology (27,28). Furthermore, various commensal bacteria have been shown to produce small molecules with anti-bacterial (29), analgesic and neuromodulatory properties (30-33). Observations such as these may well herald a new era in therapeutics: pharmabiotics (34-36).
The microbiota in the pathogenesis of gastrointestinal and liver disease
While the role of the gut microbiota in a number of disease states, ranging from enteric infections to Helicobacter pylori infection, Clostridium difficile-associated disease, small intestinal bacterial overgrowth, portalsystemic encephalopathy, spontaneous bacterial peritonitis and biliary and pancreatic sepsis, is well established, the advent of high-throughput methodologies has resulted in explorations of changes in the microbiota in a host of intestinal and liver diseases and disorders (11,12,37-41). While some of the reported associations have resulted in plausible hypotheses to support a role for the microbiota or its interactions with the host in the pathogenesis of these disorders a number of notes of caution need to be mentioned. The first of these relates to source of the microbial samples; for reasons of convenience this has usually been fecal and not mucosal and colonic rather than small intestinal or gastric. These discrepancies are not unimportant as different bacterial populations may inhabit the lumen and the juxta-mucosal surface, in the biofilm (42,43). Furthermore, while metabolic interactions between the microbiota and food or products of digestion are most likely to occur in the lumen, juxta-mucosal species and strains may be more relevant to immunological interactions (44,45). Similarly, immunological interactions are more likely to be the province of the microbiota of the small intestine, a population that, for obvious reasons, has been scarcely studied, given that this organ contains a greater mass of immune tissue than the colon. Furthermore, most studies have been based on single point in time "snapshots", have failed to account for potential instability in the microbiota over time (46) and the likely impact of diet and other environmental factors (47-49). For these and other reasons, such studies are usually incapable of correlating microbial signatures with disease activity.
However, well characterized examples of the consequences of perturbing the microbiota do exist. A vivid example is provided by antibiotic-associated diarrhea and its deadliest manifestation, Clostridium difficile colitis (50). Similar perturbations in the flora are thought to be involved in a devastating form of intestinal inflammation that may occur in new-born and, especially, premature, infants: necrotizing enterocolitis (51). In other situations, bacteria may simply be where they should not be: impaired motility and/or acid secretion from the stomach, promote an environment conducive to the proliferation, in the small intestine, of organisms normally confined to the colon and small bowel bacterial overgrowth (SIBO) ensues (52). While SIBO has traditionally been regarded as a clause of the malabsorption syndrome there has been some enthusiasm, of late, to extend the clinical manifestations of SIBO into unexplained diarrhea and, most controversially, irritable bowel syndrome (IBS) (53). In other situations, such as inflammatory bowel disease, the host-microbiota immune interaction goes ary with the host coming to recognize commensals, not as friend, but as foe and mounting an inappropriate inflammatory response (54-57). If damage to the intestinal epithelium renders the gut wall leaky and permits enteric bacteria (in whole or in part) to gain direct access to the submucosal compartments or translocate into the systemic circulation the stage is set for the development of potentially catastrophic sepsis syndromes; a scenario all too familiar to those who work in the intensive care unit (58-60). A similar scenario has been described in the aftermath of inflammatory disorders (61).
Most recently, qualitative changes in the microbiota have been invoked in the pathogenesis of a global epidemic, obesity (and its attendant consequences, the metabolic syndrome and non-alcoholic fatty liver disease (NAFLD) (17,62). It has been postulated that a shift in the composition of the flora towards a population where bacteria that are more avid extractors of absorbable nutrients delivers more calories to the host and thus contributes to obesity.
Further layers of complexity continue to be added to the host-microbiome interface. Firstly, it must be remembered that the microbiota-host interaction is a twoway conversation; it is evident that the host can also affect the composition and function of the microbiome (63).
Interventions to modify the microbiota
A variety of strategies may be employed either singly or in combination to modify the microbiota.
1. Diet
Not only is it evident that changes in diet can dramatically alter the composition of the microbiome (21,22,47-49), but it has also been postulated that consequent changes in bacterial metabolism resulting in alterations in the production of short-chain fatty acids, polysaccharide A and peptidoglycan, molecules with known immunomodulatory and immuno-regulatory functions (64), may play a role in the genesis of inflammatory disorders. Metabolic products resulting from the impact of the microbiota on dietary constituents (in this case phosphatidylcholine) have even been implicated in the pathogenesis of cardiovascular disease (65). Indeed, diet, either deliberately or inadvertently, may well represent the most common strategy given the presence of substances with prebiotic properties in a variety of food-stuffs and additives.
2. Antibiotics
Antibiotics are the most time honored approach and their impact may be complicated by unwanted changes in the commensal flora; changes that may be more extensive and long-lasting than formerly thought. One major consequence of antibiotic us is the suppression of the commensal population; while many species and strains may rebound promptly on cessation of therapy, not all do (66-69).
The use of antibiotics in inflammatory bowel disease (IBD) has witnessed somewhat of resurgence with the publication of studies illustrating benefits for antibiotic therapy in Crohns disease (70-72) and pouchitis (73) and of strategies directed against specific organisms in ulcerative colitis (74). In irritable bowel syndrome, the poorly absorbed antibiotic rifaximin has been associated with modest, though consistent, symptomatic benefit; its precise method of action remains unclear (75). Another twist to the antibiotic tale in IBD has been provided by a number of studies associating risk for IBD in later life with antibiotic exposure in early childhood (76-78); further evidence of the potential clinical impact of perturbing the microbiota at a time of its most rapid evolution (79).
The roles of antibiotic therapy in hepatic encephalopathy and spontaneous bacterial peritonitis have well documented though relationships between these effects and changes in the microbiota remain unclear. Antibiotic therapy has also been shown to reduce mortality and re-bleeding after variceal hemorrhage and its impact on liver disease, per se, has also been explored, especially, in relation to jejuno-ileal bypass-related liver disease and other liver diseases that have been linked to SIBO, translocation, bacterial antigens such as intestinal failure-related liver disease and primary sclerosing cholangitis (41). Given the evidence accumulating for a causative role for the microbiota in the various manifestations of non-alcoholic fatty liver disease (NAFLD) (40,80,81), studies of antibiotic therapy or other manipulations of the microbiota in this disease will be of especial interest.
3. Prebiotics and probiotics
Though probiotics and prebiotics have been extensively used to address a host of digestive symptoms and disorders, firm clinical data is somewhat thin on the ground (82,83). For probiotics, the best evidence for efficacy lies in diarrheal disease (84-86) and necrotizing enterocolitis (87). Studies in inflammatory bowel disease are supportive of their use in pouchitis and, possibly, in the maintenance of mild ulcerative colitis but results in Crohns disease have been disappointing (88). Though used empirically by sufferers for decades, more recently, a rationale for the use of probiotics in irritable bowel syndrome has emerged (89). While meta-analyses suggest overall efficacy for probiotics and bifidobacteria, in particular, in irritable bowel syndrome (90), it is likely that these effects, as elsewhere, are strain specific (91). There is, as yet, no convincing evidence for a role for probiotics in liver disease or its complications.
Fecal transplantation is now a widely accepted approach for the treatment of resistant, recurrent C difficile infection (92-94) and its application to other indications is being explored (95).
The future
What is the future for this area? Avenues currently under active exploration include the use of "dead bacteria, bacterial components and small molecules produced by bacteria. The possibility that bacteria could be genetically modified to transport biologically active compounds or vaccines represents another exciting application of this technology (96).
REFERENCES
1. Fraher MH, OToole PW, Quigley FM. Techniques used to characterize the gut microbiota: a guide for the clinician. Nat Rev Gastroenterol Hepatol. 2012;9(6):312-22.
2. Weinstock GM. Genomic approaches to studying the human microbiota. Nature. 2012;489(7415):250-6.
3. Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464(7285):59-65.
4. Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, et al. Enterotypes of the human gut microbiome. Nature. 2011;473(7346):174-80.
5. Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486(7402):222-7.
6. Human Microbiome Project Consortium. A framework for human microbiome research. Nature. 2012; 486(7402):215-21.
7. Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486(7402):207-14.
8. Wen L, Ley RE, Volchkov PY, Stranges PB, Avanesyan L, Stonebraker AC, et al. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature. 2008;455(7216):1109-13.
9. Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, et al. A metagenome wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490(7418):55-60.
10. Delhaes L, Monchy S, Fréalle E, Hubans C, Salleron J, Leroy S, et al. The airway microbiota in cystic fibrosis: a complex fungal and bacterial community--implications for therapeutic management. PLoS One. 2012;7(4):e36313.
11. Guarner F, Malagelada JR. Gut flora in health and disease. Lancet. 2003;361(9356):512-9.
12. Aziz Q, Doré J, Emmanuel A, Guarner F, Quigley EM. Gut microbiota and gastrointestinal health: current concepts and future directions. Neurogastroenterol Motil. 2013;25(1):4-15.
13. Lathrop SK, Bloom SM, Rao SM, Nutsch K, Lio CW, Santacruz N, et al. Peripheral education of the immune system by colonic commensal microbiota. Nature. 2011;478(7368):250-4.
14. Maynard CL, Elson CO, Hatton RD, Weaver CT. Reciprocal interactions of the intestinal microbiota and immune system. Nature. 2012;489(7415):231-41.
15. Konieczna P, Groeger D, Ziegler M, Frei R, Ferstl R, Shanahan F, et al. Bifidobacterium infantis 35624 administration induces Foxp3 T regulatory cells in human peripheral blood: potential role for myeloid and plasmacytoid dendritic cells. Gut. 2012;61(3):354-66.
16. Ley RE, Backhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci U S A. 2005;102(31):11070-5.
17. Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444(7122):1022-3.
18. Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444(7122):1027-31.
19. Turnbaugh PJ, Backhed F, Fulton L, Gordon JI. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe. 2008;3(4):213-23.
20. Tremaroli V, Bäckhed F. Functional interactions between the gut microbiota and host metabolism. Nature. 2012;489(7415):242-9.
21. Murphy EF, Cotter PD, Healy S, Marques TM, OSullivan O, Fouhy F, et al. Composition and energy harvesting capacity of the gut microbiota: relationship to diet, obesity and time in mouse models. Gut. 2010;59(12):1635-42.
22. Murphy EF, Cotter PD, Hogan A, OSullivan O, Joyce A, Fouhy F, et al. Divergent metabolic outcomes arising from targeted manipulation of the gut microbiota in diet-induced obesity. Gut. 2013;62(2):220-6.
23. Wall R, Ross RP, Shanahan F, OMahony L, OMahony C, Coakley M, et al. Metabolic activity of the enteric microbiota influences the fatty acid composition of murine and porcine liver and adipose tissues. Am J Clin Nutr. 2009;89(5):1393-401.
24. Wall R, Ross RP, Shanahan F, OMahony L, Kiely B, Quigley E, et al. Impact of administered bifidobacterium on murine host fatty acid composition. Lipids. 2010;45(5):429-36.
25. Rosberg-Cody E, Stanton C, OMahony L, Wall R, Shanahan F, Quigley EM, et al. Recombinant lactobacilli expressing linoleic acid isomerase can modulate the fatty acid composition of host adipose tissue in mice. Microbiology. 2011;157(Pt 2):609-15.
26. Wall R, Marques TM, OSullivan O, Ross RP, Shanahan F, Quigley EM, et al. Contrasting effects of Bifidobacterium breve NCIMB 702258 and Bifidobacterium breve DPC 6330 on the composition of murine brain fatty acids and gut microbiota. Am J Clin Nutr. 2012;95(5):1278-87.
27. Barrett E, Fitzgerald P, Dinan TG, Cryan JF, Ross RP, Quigley EM, et al. Bifidobacterium breve with α-linolenic acid and linoleic acid alters fatty acid metabolism in the maternal separation model of irritable bowel syndrome. PLoS One. 2012;7(11):e48159.
28. Heijtz RD, Wang S, Anuar F, Qian Y, Björkholm B, Samuelsson A, et al. Normal gut microbiota modulates brain development and behavior. Proc Natl Acad Sci U S A. 2011;108(7):3047-52.
29. Bravo JA, Forsythe P, Chew MV, Escaravage E, Savignac HM, Dinan TG, et al. Ingestion of Lactobacillus strain regulates emotional behavior and central GABA receptor expression in a mouse via the vagus nerve. Proc Natl Acad Sci U S A. 2011;108(38):16050-5.
30. Rea MC, Sit CS, Clayton E, OConnor PM, Whittal RM, Zheng J, et al. Thuricin CD, a posttranslationally modified bacteriocin with a narrow spectrum of activity against Clostridium difficile. Proc Natl Acad Sci U S A. 2010;107(20):9352-7.
31. Rousseaux C, Thuru X, Gelot A, Barnich N, Neut C, Dubuquoy L, et al. Lactobacillus acidophilus modulates intestinal pain and induces opioid and cannabinoid receptors. Nature Med. 2007;13(1):35-7.
32. Verdu EF, Bercik P, Bergonzelli GE, Huang XX, Blenerhasset P, Rochat F, et al. Lactobacillus paracasei normalizes muscle hypercontractility in a murine model of postinfective gut dysfunction. Gastroenterology. 2004;127(3):826-37.
33. Verdu EF, Bercik P, Verma-Gandhu M, Huang XX, Blenerhasset P, Jackson W, et al. Specific probiotic therapy attenuates antibiotic induced visceral hypersensitivity in mice. Gut. 2006;55(2):182-90.
34. Kamiya T, Wang L, Forsythe P, Goettsche G, Mao Y, Wang Y, et al. Inhibitory effects of Lactobacillus reuteri on visceral pain induced by colorectal distension in Sprague-Dawley rats. Gut. 2006;55(2):191-6.
35. Sleator RD, Hill C. Engineered pharmabiotics with improved therapeutic potential. Hum Vaccin. 2008;4(4):271-4.
36. Shanahan F, Dinan TG, Ross P, Hill C. Probiotics in transition. Clin Gastroenterol Hepatol. 2012;10(11):1220-4.
37. Ghoshal UC, Shukla R, Ghoshal U, Gwee KA, Ng SC, Quigley EM. The gut microbiota and irritable bowel syndrome: friend or foe? Int J Inflam. 2012;2012:151085.
38. Jeffery IB, Quigley EM, Öhman L, Simrén M, OToole PW. The microbiota link to irritable bowel syndrome: an emerging story. Gut Microbes. 2012;3(6):572-6.
39. Quigley EM. The enteric microbiota in the pathogenesis and management of constipation. Best Pract Res Clin Gastroenterol. 2011;25(1):119-26.
40. Abu-Shanab A, Quigley EM. The role of the gut microbiota in nonalcoholic fatty liver disease. Nat Rev Gastroenterol Hepatol. 2010;7(12):691-701.
41. Quigley EM, Stanton C, Murphy EF. The gut microbiota and the liver. Pathophysiological and clinical implications. J Hepatol. 2013;58(5):1020-7.
42. Codling C, OMahony L, Shanahan F, Quigley EM, Marchesi JR. A molecular analysis of fecal and mucosal bacterial communities in irritable bowel syndrome. Dig Dis Sci. 2010;55(2):392-7.
43. Carroll IM, Ringel-Kulka T, Keku TO, Chang YH, Packey CD, Sartor RB, et al. Molecular analysis of the luminal-and mucosal associated intestinal microbiota in diarrhea-predominant irritable bowel syndrome. Am J Physiol Gastrointest Liver Physiol. 2011;301(5):G799-807.
44. Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336(6086):1268-73.
45. Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453(7195):620-5.
46. Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489(7415):220-30.
47. Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334(6052):105-8.
48. Kau AL, Ahern PP, Griffin NW, Goodman AL, Gordon JI. Human nutrition, the gut microbiome and the immune system. Nature. 2011;474(7351):327-36.
49. Claesson MJ, Jeffery IB, Conde S, Power SE, OConnor EM, Cusack S, et al. Gut microbiota composition correlates with diet and health in the elderly. Nature. 2012;488(7410):178-84.
50. Kelly CP, LaMont JT. Clostridium difficile--more difficult than ever. N Engl J Med. 2008;359(18):1932-40.
51. Neu J, Mshvildadze M, Mai V. A roadmap for understanding and preventing necrotizing enterocolitis. Curr Gastroenterol Rep. 2008;10(5):450-7.
52. Quigley EM, Abu-Shanab A. Small intestinal bacterial overgrowth. Infect Dis Clin North Am. 2010;24(4):943-59.
53. Vanner S. The small bowel intestinal bacterial overgrowth. Irritable bowel syndrome hypothesis: implications for treatment. Gut. 2008;57(9):1315-21.
54. Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A. 2007;104(34):13780-5.
55. Sartor RB. Genetics and environmental interactions shape the intestinal microbiome to promote inflammatory bowel disease versus mucosal homeostasis. Gastroenterology. 2010;139(6):1816-9.
56. Willing BP, Dicksved J, Halfvarson J, Andersson AF, Lucio M, Zheng Z, et al. A pyrosequencing study in twins shows that gastrointestinal microbial profiles vary with inflammatory bowel disease phenotypes. Gastroenterology. 2010;139(6):1844-54.
57. Shanahan F. The microbiota in inflammatory bowel disease: friend, bystander, and sometime-villain. Nutr Rev. 2012;70 Suppl 1:S31-7.
58. De Smet AM, Kluytmans JA, Cooper BS, Mascini EM, Benus RF, van der Werf TS, et al. Decontamination of the digestive tract and oropharynx in ICU patients. N Engl J Med. 2009;360(1):20-31.
59. Gatt M, Reddy BS, MacFie J. Review article: bacterial translocation in the critically ill – evidence and prevention. Aliment Pharmacol Ther. 2007;25(7):741-57.
60. Quigley EM. Passing the bug-Translocation, bacteremia, and sepsis in the intensive care unit patient: Is intestinal decontaminationtheanswer? CritCareMed.2011;39(5):1202-3.
61. Asfaha S, Macnaughton WK, Appleyard CB, Chadee K, Wallace JL. Persistent epithelial dysfunction and bacterial translocation after resolution of intestinal inflammation. Am J Physiol Gastrointest Liver Physiol. 2001;281(3):G635-44.
62. Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, et al. A core guts microbiome in obese and lean twins. Nature. 2009;457(7228):480-4.
63. Kotarsky K, Sitnik KM, Stentsad H, Kotarsky H, Schmidtchen A, Koslowski M, et al. A novel role for constitutively expressed epithelial-derived chemokines as antibacterial peptides in the intestinal mucosa. Mucosal Immunol. 2010;3(1):40-8.
64. Maslowski KM, Mackay CR. Diet, gut microbiota and immune function. Nature Immunol. 2011;12(1):5-9.
65. Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, Dugar B, et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472(7341):57-63.
66. Jernberg C, Löfmark S, Edlund C, Jansson JK. Long-term impacts of antibiotic exposure on the human intestinal microbiota. Microbiology. 2010;156(Pt 11):3216-23.
67. Dethlefsen L, Relman DA. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci U S A. 2011;108 Suppl 1:4554-61.
68. Cotter PD, Stanton C, Ross RP, Hill C. The impact of antibiotics on the gut microbiota as revealed by high throughput DNA sequencing. Discov Med. 2012;13(70):193-9.
69. OSullivan O, Coakley M, Lakshminarayanan B, Conde S, Claesson MJ, Cusack S, et al. Alterations in intestinal microbiota of elderly Irish subjects post-antibiotic therapy. J Antimicrob Chemother. 2013;68(1):214-21.
70. Feller M, Huwiler K, Schoepfer A, Shang A, Furrer H, Egger M. Long-term antibiotic treatment for Crohns disease: systematic review and meta-analysis of placebo-controlled trials. Clin Infect Dis. 2010;50(4):473-80.
71. Doherty GA, Bennett GC, Cheifetz AS, Moss AC. Meta analysis: targeting the intestinal microbiota in prophylaxis for post-operative Crohns disease. Aliment Pharmacol Ther. 2010;31(8):802-9.
72. Prantera C, Lochs H, Grimaldi M, Danese S, Scribano ML, Gionchetti P, et al. Rifaximin-extended intestinal release induces remission in patients with moderately active Crohns disease. Gastroenterology. 2012;142(3):473-81.
73. Holubar SD, Cima RR, Sandborn WJ, Pardi DS. Treatment and prevention of pouchitis after ileal pouch-anal anastomosis for chronic ulcerative colitis. Cochrane Database Syst Rev. 2010;(6):CD001176.
74. Ohkusa T, Kato K, Terao S, Chiba T, Mabe K, Murakami K, et al. Newly developed antibiotic combination therapy for ulcerative colitis: a double-blind placebo-controlled multicenter trial. Am J Gastroenterol. 2010;105(8):1820-9.
75. Pimentel M, Lembo A, Chey WD, Zakko S, Ringel Y, Yu J, et al. Rifaximin therapy for patients with irritable bowel syndrome without constipation. N Engl J Med. 2011;364(1):22-32.
76. Shaw SY, Blanchard JF, Bernstein CN. Association between the use of antibiotics in the first year of life and pediatric inflammatory bowel disease. Am J Gastroenterol. 2010;105(12):2687-92.
77. Shaw SY, Blanchard JF, Bernstein CN. Association between the use of antibiotics and new diagnoses of Crohns disease and ulcerative colitis. Am J Gastroenterol. 2011;106(12):2133-42.
78. Shaw SY, Blanchard JF, Bernstein CN. Association between early childhood otitis media and pediatric inflammatory bowel disease: an exploratory population-based analysis. J Pediatr. 2013;162(3):510-4.
79. Cho I, Yamanishi S, Cox L, Methé BA, Zavadil J, Li K, et al. Antibiotics in early life alter the murine colonic microbiome and adiposity. Nature. 2012;488(7413):621-6.
80. Henao-Mejia J, Elinav E, Jin C, Hao L, Mehal WZ, Strowig T, et al. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature. 2012;482(7384):179-85.
81. Raman M, Ahmed I, Gillevet PM, Probert CS, Ratcliffe NM, Smith S, et al. Fecal Microbiome and Volatile Organic Compound Metabolome in Obese Humans with Nonalcoholic Fatty Liver Disease. Clin Gastroenterol Hepatol. 2013 [epub ahead of print]
82. Quigley EM. Gut microbiota and the role of probiotics in therapy. Curr Opin Pharmacol. 2011;11(6):593-603.
83. Sanders ME, Guarner F, Guerrant R, Holt PR, Quigley EM, Sartor RB, et al. An update on the use and investigation of probiotics in health and disease. Gut. 2013;62(5):787-96.
84. NASPGHAN Nutrition Report Committee; Michail S, Sylvester F, Fuchs G, Issenman R. Clinical efficacy of probiotics: review of the evidence with focus on children. J Pediatr Gastroenterol Nutr. 2006;43(4):550-7.
85. Floch MH, Walker WA, Guandalini S, Hibberd P, Gorbach S, Surawicz C, et al. Recommendations for probiotic use--2008. J Clin Gastroenterol. 2008;42 Suppl 2:S104-8.
86. Preidis GA, Hill C, Guerrant RL, Ramakrishna BS, Tannock GW, Versalovic J. Probiotics, enteric and diarrheal diseases, and global health. Gastroenterology. 2011;140(1):8-14.
87. Lin HC, Hsu CH, Chen HL, Chung MY, Hsu JF, Lien RI, et al. Oral probiotics prevent necrotizing enterocolitis in very low birth weight preterm infants: a multicenter, randomized, controlled trial. Pediatrics. 2008;122(4):693-700.
88. Veerappan GR, Betteridge J, Young PE. Probiotics for the treatment of inflammatory bowel disease. Curr Gastroenterol Rep. 2012;14(4):324-33.
89. Quigley FM, Flourie B. Probiotics in irritable bowel syndrome: a rationale for their use and an assessment of the evidence to date. Neurogastroenterol Motil. 2007;19(3):166-72.
90. Moayyedi P, Ford AC, Talley NJ, Cremonini F, Foxx-Orenstein A, Brandt L, et al. The efficacy of probiotics in the therapy of irritable bowel syndrome: a systematic review. Gut. 2010;59(3):325-32.
91. Brenner DM, Moeller MJ, Chey WD, Schoenfeld PS. The utility of probiotics in the treatment of irritable bowel syndrome: a systematic review. Am J Gastroenterol. 2009;104(4):1033-49.
92. van Nood E, Vrieze A, Nieuwdorp M, Fuentes S, Zoetendal EG, de Vos WM, et al. Duodenal infusion of donor feces for recurrent Clostridium difficile. N Engl J Med. 2013;368(5):407-15.
93. Guo B, Harstall C, Louie T, Veldhuyzen van Zanten S, Dieleman LA. Systematic review: faecal transplantation for the treatment of Clostridium difficile-associated disease. Aliment Pharmacol Ther. 2012;35(8):865-75.
94. Kassam Z, Lee CH, Yuan Y, Hunt RH. Fecal Microbiota Transplantation for Clostridium difficile Infection: Systematic Review and Meta-Analysis. Am J Gastroenterol. 2013;108(4):500-8.
95. Vrieze A, Van Nood E, Holleman F, Salojärvi J, Kootte RS, Bartelsman JF, et al. Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology. 2012;143(4):913-6.
96. Shanahan F, Collins SM. Pharmabiotic manipulation of the microbiota in gastrointestinal disorders, from rationale to reality. Gastroenterol Clin North Am. 2010;39(3):721-6.
Citar como: Quigley EMM, Monsour HP. Targeting the microbiota in the management of gastrointestinal and liver disease. Rev Gatroenterol Peru. 2013;33(2):139-44.
Correspondence:
Eamonn M M Quigley MD
6550 Fannin St. Suite SM 1001
Houston, Texas 77030, USA
E-mail: equigley@tmhs.org
Recibido: 15/04/2013
Aprobado: 03/05/2013