Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO
Related links
-
 Similars in
SciELO
Similars in
SciELO
Share
Revista Peruana de Medicina Experimental y Salud Publica
Print version ISSN 1726-4634
Rev. perú. med. exp. salud publica vol.39 no.1 Lima Jan./Mar. 2022 Epub Mar 17, 2022
http://dx.doi.org/10.17843/rpmesp.2022.391.10861
Letters to Editor
Emergence of the Cosmopolitan genotype of dengue virus serotype 2 (DENV2) in Madre de Dios, Peru, 2019
1 Laboratorio Nacional de Referencia de Metaxénicas Virales, Centro Nacional de Salud Pública, Instituto Nacional de Salud, Lima, Peru.
2 Laboratorio Nacional de Referencia de Biología Molecular y Biotecnología, Centro Nacional de Salud Pública, Instituto Nacional de Salud, Lima, Peru.
3 Dirección Ejecutiva de Epidemiología, Dirección Regional de Salud de Madre de Dios, Madre de Dios, Peru.
4 Centro Nacional de Salud Pública, Instituto Nacional de Salud, Lima, Peru.
To the editor. Dengue is a viral disease transmitted by Aedes aegypti that is increasingly spreading in tropical and subtropical areas. Up to the epidemiological week 52 (December) of 2019, 3,139,335 cases of dengue were reported in the Americas with deaths (lethality 0.049%); while in Peru during that same epidemiological week, 15,297cases of dengue were reported, of which 76.0% corresponded to the Madre de Dios, Loreto, San Martin, Cajamarca and Tumbes regions. In the Madre de Dios region, 4893 cases were reported, 4.3 times more than in 2018. Of the 13,708 cases in these regions, 3831 (78.3%) showed no alarm signs and 1020 (20.8%) did show alarm signs; 42 (0.86%) were severe and 18 (0.37%) died 1.
Dengue virus (DENV) comprises four serotypes, and each serotype is further divided into distinct genotypes. The increase in the incidence of dengue and the severity of the disease have been associated with the change in circulating genotypes as well as viral evolution.
Serotypes DENV1, DENV2, DENV3, and DENV4 have circulated since 2014 in the Madre de Dios region, located in the Peruvian Amazon 2. The aim of this study was to show the introduction of a new DENV2 genotype, which does not circulate in the Americas.
During the 2019 dengue outbreak in Madre de Dios, initial diagnosis was made in patients by detecting the NS1 antigen with ELISA. Viral isolation in the C6-36 cell line was successful in 12 samples out of 32 processed. The virus was identified with monoclonal antibodies by indirect immunofluorescence, and simultaneously the samples were analyzed by real-time RT-PCR (qPCR-RT) at the National Referrral Laboratory of Viral Metaxenics of the Instituto Nacional de Salud (INS). All samples were identified as DENV2.
Following the protocol of Santiago et al. 3, the amplified and purified products were sequenced by the Sanger method at the INS Molecular Biology Laboratory. The obtained sequences were assembled using the SeqScape software to obtain the complete consensus sequence of the E gene. The complete gene sequences were used for phylogenetic analysis using the MEGA X software and the Maximum Likehook algorithm. In addition, the support of each clade was evaluated using the Bootstrap method.
Phylogenetic analysis of twelve evaluated samples groups the sequences into two known DENV2 genotypes; two samples from Madre de Dios cluster in the America/Asia genotype clade that has been circulating in the Loreto region since late 2010 and has been widely distributed since then in Peru. The other ten samples cluster in the Cosmopolitan genotype clade, together with strains from Bangladesh 4 and China 5, as shown in the phylogenetic tree (Figure 1).
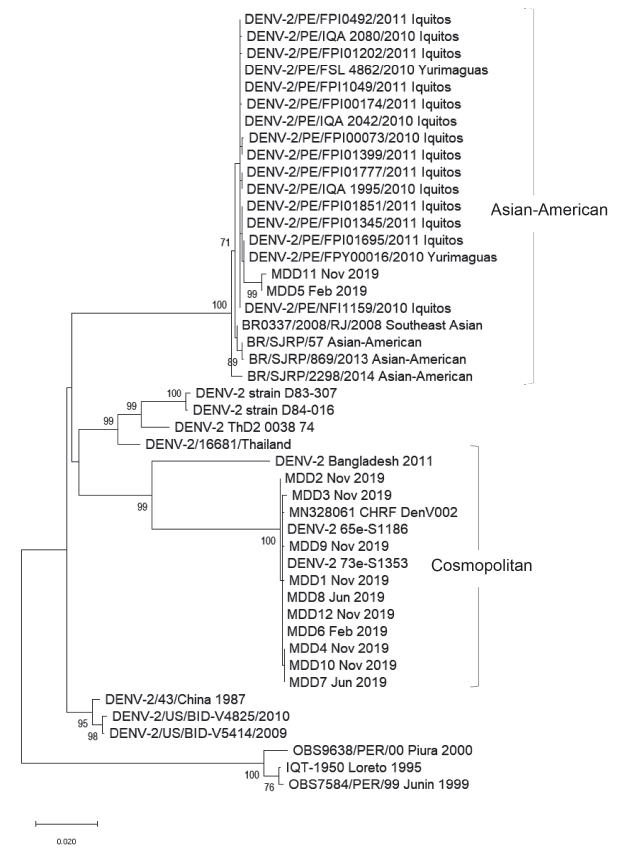
Figure 1 Phylogenetic tree constructed with the Maximum Likehook algorithm showing the homology of DENV2 with the Cosmopolitan genotype from the Madre de Dios-Peru outbreak, 2019. Bootstrap value (>70) is represented at the root of each cluster.
These results demonstrate the introduction of a “new genotype” of DENV2 derived from the Cosmopolitan genotype, highly homologous to the strains reported in Bangladesh. These results were confirmed by the Centers for Disease Control and Prevention, Dengue Branch-Puerto Rico. This Cosmopolitan genotype is circulating in other parts of the world such as India 4, Africa 6, China 5, Indian Ocean and recently (2019) in the French Polynesia 7. To our knowledge, this is the first report of an outbreak caused by a Cosmopolitan genotype in the Americas region. The first report of a Cosmopolitan strain detected in the Americas was described in the late 1990s in the Yucatan Peninsula in Mexico, but only one viral isolate was studied and so far, this genotype has not been detected in the Americas region. Determination of the new genotype was made from viral isolates and retrospectively there were limitations to have detailed clinical information.
This finding highlights the need to activate routine genomic surveillance in the laboratory as well as the need for it to include clinical-epidemiological information. It is necessary to conduct genomic surveillance of DENV 2 isolates from other endemic areas of the country to have a broader view of the circulation and to evaluate the route of entry and the spread of this new genotype in the Americas. Additionally, the study should be complemented in order to determine whether or not this new genotype is associated with severe cases of dengue.
Acknowledgements:
To the staff of the Refrerral Laboratory of Madre de Dios and to Biologist Edwin Tineo for handling the samples; to the staff of the National Referral Laboratory of Viral Metaxenic Diseases and to Biologist Adolfo Marcelo and Lic. Nancy Susy Merino for processing the samples; to biologists Omar Cáceres and Henry Baylón of the National Referral Laboratory of Molecular Biology for their support with the phylogenetic analysis and to Dr. Oscar Escalante of the National Public Health Center of the Instituto Nacional de Salud for his opinion on the phylogenetic analysis; to Dr. Karim Pardo, executive director of DEPCEM-MINSA and Dr. Manuel Espinoza for their support with the clinical part of the outbreak intervention. To biologist Fernando Chapilliquen of the Centro Nacional de Epidemiología Prevención y Control de Enfermedades -CDC-Perú, for his epidemiological contributions. Special thanks to Dr. Leticia Franco and Dr. Jairo Mendez for their opinion on the phylogenetic analysis and for linking us with the Dengue Genomic Surveillance (ViGenDA) project of the Arbovirus Laboratory Network (RELDA) of PAHO and to Dr. Gilberto Santiago and Dr. Jorge Muñoz of the Centers for Disease Control and Prevention, Dengue Branch - Puerto Rico, for confirming the results.
REFERENCES
. Ministerio de Salud. Sala de situación virtual, Dengue. [Internet]. Lima: Centro Nacional de Epidemiología y control de Enfermedades, MINSA; 2019 [Citado el 5 de diciembre 2019]. Disponible en: https://www.dge.gob.pe/portal/index.php?option=com_content&view=article&i- d=14&Itemid=154. [ Links ]
. Cabezas C, Fiestas V, García-Mendoza M, Palomino M, Mamani E, Donaires F. Dengue en el Perú: a un cuarto de siglo de su reemergencia. Rev Peru Med Exp Salud Publica. 2015;32(1):146-56. [ Links ]
. Santiago GA, González GL, Cruz-López F, Muñoz-Jordan JL. Development of a standardized Sanger-based method for partial sequencing and genotyping of dengue viruses. J Clin Microbiol. 2019 Mar 28;57(4):e01957-18. doi: 10.1128/JCM.01957-18. [ Links ]
. Suzuki K, Phadungsombat J, Nakayama EE, Saito A, Egawa A, Sato T, et al. Genotype replacement of dengue virus type 3 and clade replace-ment of dengue virus type 2 genotype Cosmopolitan in Dhaka, Bangladesh in 2017. Infect Genet Evol. 2019;75:103977. doi: 10.1016/j.meegid.2019.103977. [ Links ]
. Yenamandra SP, Koo C, Chiang S, Lim HSJ, Yeo ZY, Ng LC, et al. Evolution, heterogeneity and global dispersal of cosmopolitan genotype of Dengue virus type 2. Sci Rep. 2021;11(1):13496. doi: 10.1038/s41598-021-92783-y. [ Links ]
. Hill SC, Neto de Vasconcelos J, Granja B, Thézé J, Jandondo D, Neto Z, et al. Early Genomic Detection of Cosmopolitan Genotype of Dengue Virus Serotype 2, Angola, 2018. Emerg Infect Dis. 2019;25(4):784-787. doi: 10.3201/eid2504.180958. [ Links ]
. Aubry M, Mapotoeke M, Teissier A, Paoaafaite T, Dumas-Chastang E, Giard M, et al. Dengue virus serotype 2 (DENV-2) outbreak, French Polynesia, 2019. Euro Surveill. 2019;24(29):1900407. doi: 10.2807/1560-7917.ES.2019.24.29.1900407. [ Links ]
Received: February 21, 2022; Accepted: March 09, 2022










 text in
text in