Introduction
Psychrotolerant bacterium A. ferrivorans has been successfully characterized due to its ability to oxidize Fe2+ more efficiently than A. ferrooxidans at 5 °C (Kupka et al. 2007; Escobar et al. 2010; Hallberg et al. 2010; Ccorahua-Santo et al. 2017). Compared to other Acidithiobacillus sp. strains, A. ferrivorans grows more quickly at 5 °C (t >50 h) in a standard 9K medium (33.3% Fe2+) and in the presence of other substrates such as sulphides (Ccorahua-Santo et al. 2017) and pyrite (Barahona et al. 2014). Moreover, the microorganism’s capacity to fix atmospheric carbon dioxide and its tolerance to heavy metals makes it the best candidate for industrial-scale bioleaching in extremely hostile environments, even at temperatures below 10°C. Recently, we reported the strain PQ33 of A. ferrivorans, which performs better at oxidizing ferrous iron at pH 1.6 and copper sulphide at low temperatures than other A. ferrivorans strains: A. ferrivorans SS3 (Kupka et al. 2007; Liljeqvist et al. 2011), CF27 (Talla et al. 2014) and ACH (Barahona et al. 2014). Additionally, subsequent genome and gene expression analyses have determined the principal traits of evolutionary fitness in A. ferrivorans for adaptation to low temperatures and oxidative capacity (Liljeqvist et al. 2015; Christel et al. 2016; Ccorahua-Santo et al. 2017). However, there is only one report of other genomic traits related to elements outside of the chromosomal genome (Tran et al. 2017).
On the other hand, other strains from the Acidithiobacillus genus, such as A. caldus and A. ferrooxidans, have shown traits related to functional plasmids, and some gene sequences involved in the conjugation mechanism have been found (Rohrer and Rawlings 1992; Gardner et al. 2001; Rawlings and Tietze 2001; You et al. 2011). Besides, A. ferrivorans has not been evaluated for any other traits than oxidative capacity or cold tolerance. Genome sequencing revealed that A. caldus SM-1 has one chromosome and four plasmids: pLAtc1, pLAtc2, pLAtc3 and pLAtcm (You et al. 2011). Preliminary analysis of these plasmids indicated that pLAtc1 is probably a member of the IncQ-like broad-host-range plasmid group. The IncQ-like plasmids have similar replicons comprising three essential replication genes (repA, repB, and repC) and an oriV region (Rawlings and Tietze 2001; Zhang et al. 2014).
Additionally, strains of Acidithiobacillus sp. have been shown to contain plasmids with complementary functions for adaptation in a bioleaching community. The main traits of these plasmids involve conjugation for A. caldus, and conjugation and additional heavy metal resistance genes for A. ferrooxidans and A. thiooxidans. Holmes et al. (Holmes et al. 1984) reported the mobile pTF1 plasmid (6.7 Kbp length) from A. ferrooxidans, with a 2.8 kbp region related to the mobilization function (Drolet et al. 1990). The coding sequence for the MobL protein is contained within this region and is essential for mobilization.
Furthermore, pTF-FC2 plasmid (12.2 Kbp length) from A. ferrooxidans was reported as a highly mobile plasmid (Rawlings and Kusano 1994). It comprises three regions: a replication region, a mobilization region, and a transpositional element (Rawlings 2005). Important roles have been described in relation to plasmids for species of Acidithiobacillus; however, only one plasmid has been previously isolated from A. ferrivorans (Tran et al. 2017). Despite the presence of characteristics related to plasmid stabilization in the genomes of the six A. ferrivorans strains, there are no experimental reports about the presence of plasmids in this species.
Despite the increased interest raised on A. ferrivorans, there are few studies on the role of the plasmids in the genomic context of this psychrotolerant specie. Here, we report the molecular characterization and comparative genomics of two novel plasmids (pAfPQ33-1 and pAfPQ33-2) of the recently reported A. ferrivorans PQ33 strain (Ccorahua-Santo et al. 2017). After gene annotation for those of genes encoded in two plasmids from this strain, we performed a genome-scale comparison with other plasmids from Acidithiobacillus genus. The comparison suggests that pAfPQ33-1 plasmid could be involved in the conjugation process and unveils details of its role in an evolutionary context. In addition, an origin of replication was found in both plasmids. The presence of a toxin-antitoxin system, as well as the detection of a putative VapD protein-encoding gene, suggest mechanisms for both positive and negative effects, respectively, on horizontal gene transfer in the PQ33 strain.
Materials and methods
Cell culture and harvesting
A sample of A. ferrivorans PQ33 was obtained from the Laboratory of Microbiology and Molecular Biotechnology at Universidad Nacional Mayor de San Marcos, Lima, Peru (CcorahuaSanto et al. 2017). The PQ33 strain was inoculated in 9K medium consisting of 3.33% w/v FeSO .7H O, 0.04% w/v MgSO .7H O, 0.01% w/v (NH ) SO , and 0.004% 4 2 4 2 4 w/v KH PO pH 2.0 (Ramírez et al. 2004), and incubated at room temperature (~24 °C) in a shaker at 200 rpm. The cultures that reached the exponential phase (3 5 x 108 cell/mL) in 9k medium were inoculated again in the same medium until they reached the exponential phase. The cell count was verified with a Petroff-Housser counting chamber. After the second culture, the cells were centrifuged at 13000 rpm, and the pellet was washed with acid water (pH 2.0) and centrifuged at 13000 rpm. The process was repeated as many times as necessary until the jarosite precipitated was eliminated. Then, the pellet was washed five times with 10 mM sodium citrate at pH 7.0.
Plasmid isolation
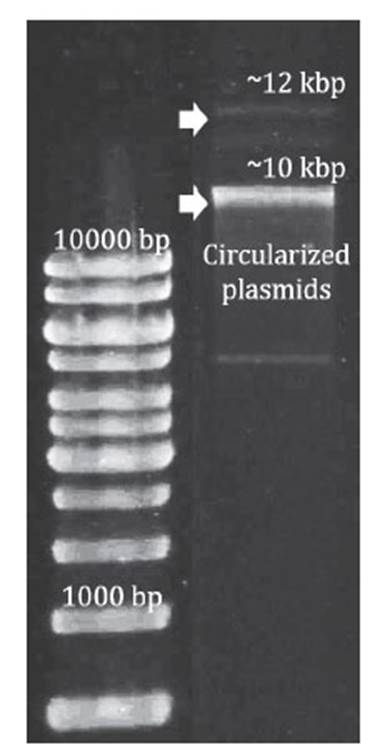
Plasmids of both 12 kbp and 10 kbp were isolated from A. ferrivorans PQ33 using the GeneJETTM Plasmid Miniprep kit following the manufacturer’s instructions. To verify plasmid size, the isolated plasmids were run on a 1% agarose gel. DNA electrophoresis was performed at 50 V for 90 minutes with a Bio-Rad® PowerPac HC. Total purified plasmid DNA was used for Illumina HiSeq sequencing technology.
Sequencing, annotation and analysis
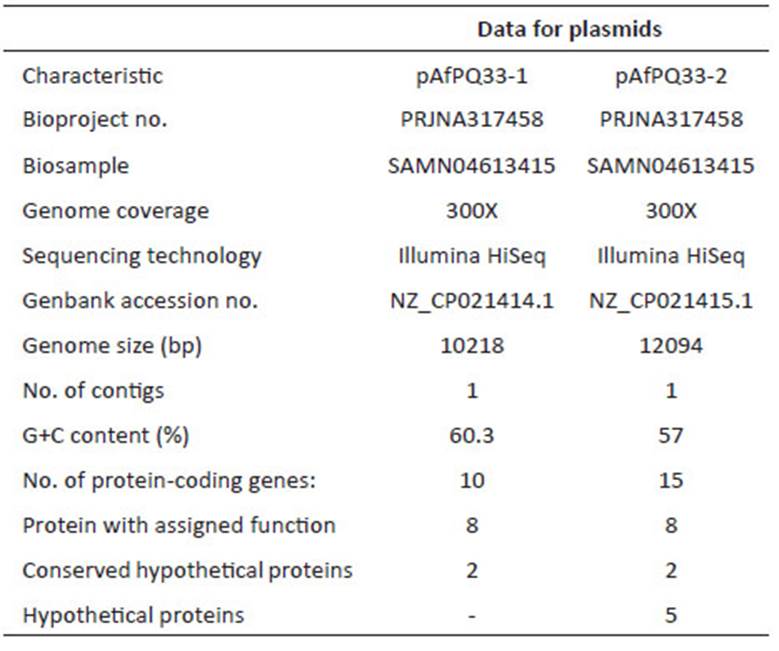
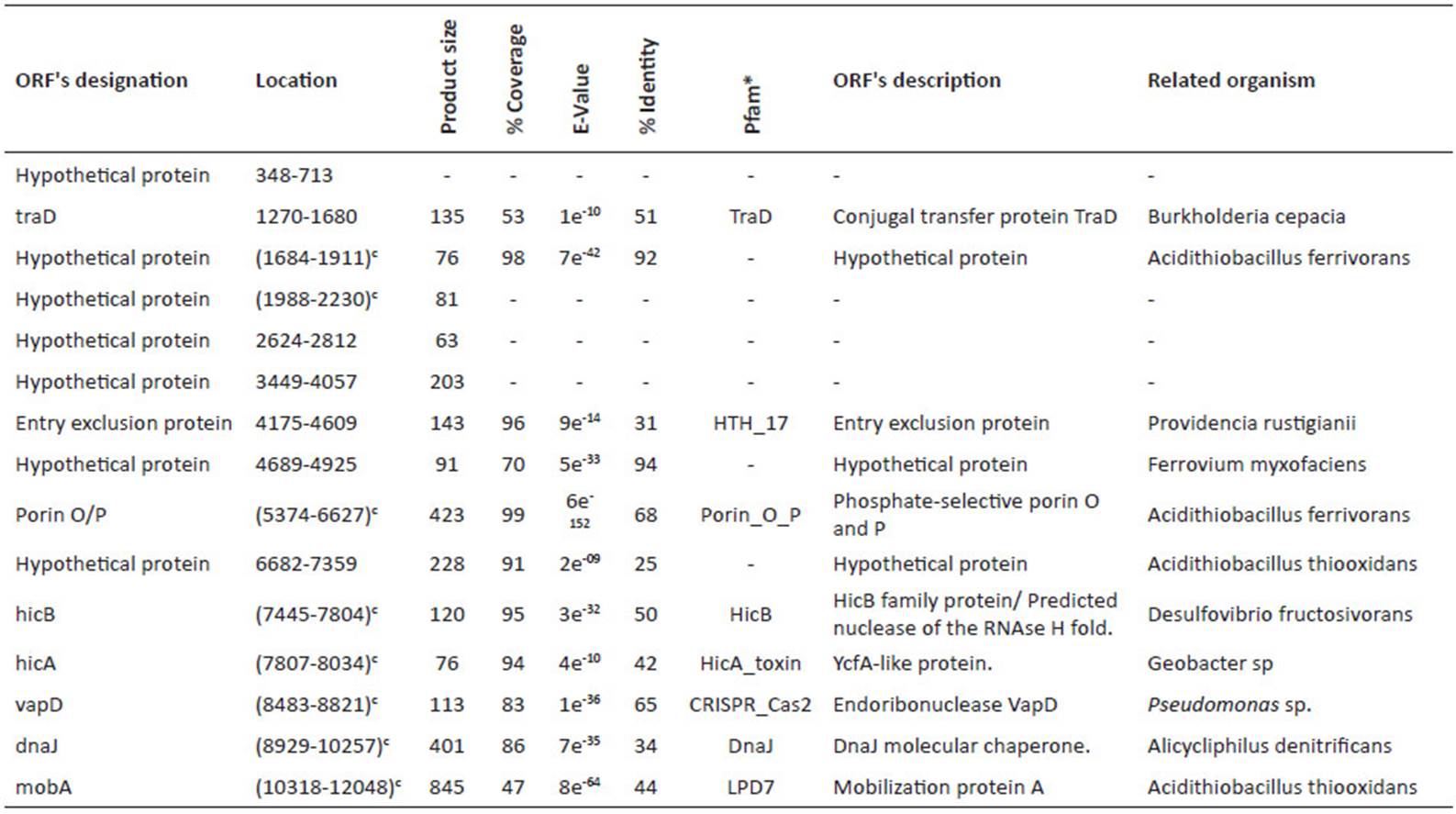
For sequencing, the library was prepared following the Illumina® TruSeqTM protocol, with an insert size of 400 bp and a size of 100 bp for the reads. The next-generation sequencing was performed using Illumina® HiSeq 2000 technology. The quality of sequencing reads was verified with FastQC software (Babraham Institute, Bioinformatics). Two de-novo assemblies were performed with Velvet 1.1 (Zerbino 2010) and SPAdes 3.07 softwares (Bankevich et al. 2012), and the best assembly was chosen. Preliminary gene annotations were performed with the Prokka tool (Seemann 2014) and ORFfinder. Within Prokka, Prodigal was run for coding sequence prediction (Hyatt et al. 2010), and the annotation was conducted by BLAST+ (Camacho et al. 2009), which was used locally against the UniProt database (Apweiler et al. 2004). For annotation, the best match was chosen. Additionally, Prokka was run with Markov Hidden Model on Hmmscan (Eddy 2011) against the Pfam (Finn et al. 2014) and TIGRFAMs databases (Haft et al. 2013). Moreover, the TRNAScanSE (Lowe and Eddy 1997) and RNAmmer (Lagesen et al. 2007) tools were used for tRNA and rRNA prediction, respectively. For assignment of putative genes, an identity percentage greater than 30% and an E-value below e-05 were required. Final annotation was performed by NCBI’s Prokaryotic Genome Automatic Annotation Pipeline (PGAP) service. The NCBI service determined 9 CDS and 13 CDS for the pAfPQ33-1 and pAfPQ33-2 plasmids, respectively. However, the cured annotation of this study was considered. The NCBI accession numbers for the pAfPQ33-1 and pAfPQ33-2 plasmids are NZ_CP021414.1 and NZ_CP021415.1, respectively (Table 1). Results of the identity and conserved pattern of the proteins related to other organisms are shown in Table 2 and Table 4.
The origin of replication was verified by the deviation of GC skew in DNAPlotter (Carver et al. 2009). Additionally, the plasmids’ origin of replication was compared with the origin of other available plasmids from Acidithiobacillus genus. The circular map and gene position in the pAFPQ33-1 and pAFPQ33-2 plasmids were generated by DNAPlotter (Carver et al. 2009). The predicted domain organization of porins O and P and CDG-PDE was performed using Pfam.
Table 2 Identified ORFs in pAfPQ33-1 plasmid of A. ferrivorans.
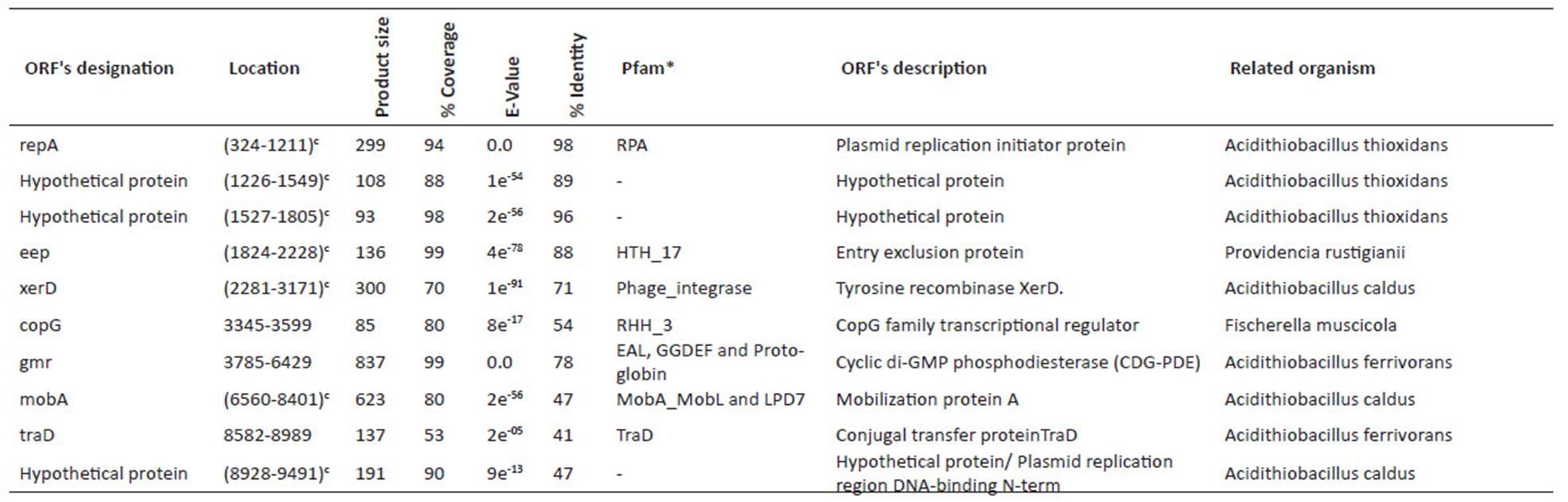
*most significant match
cGene in complementary sequence
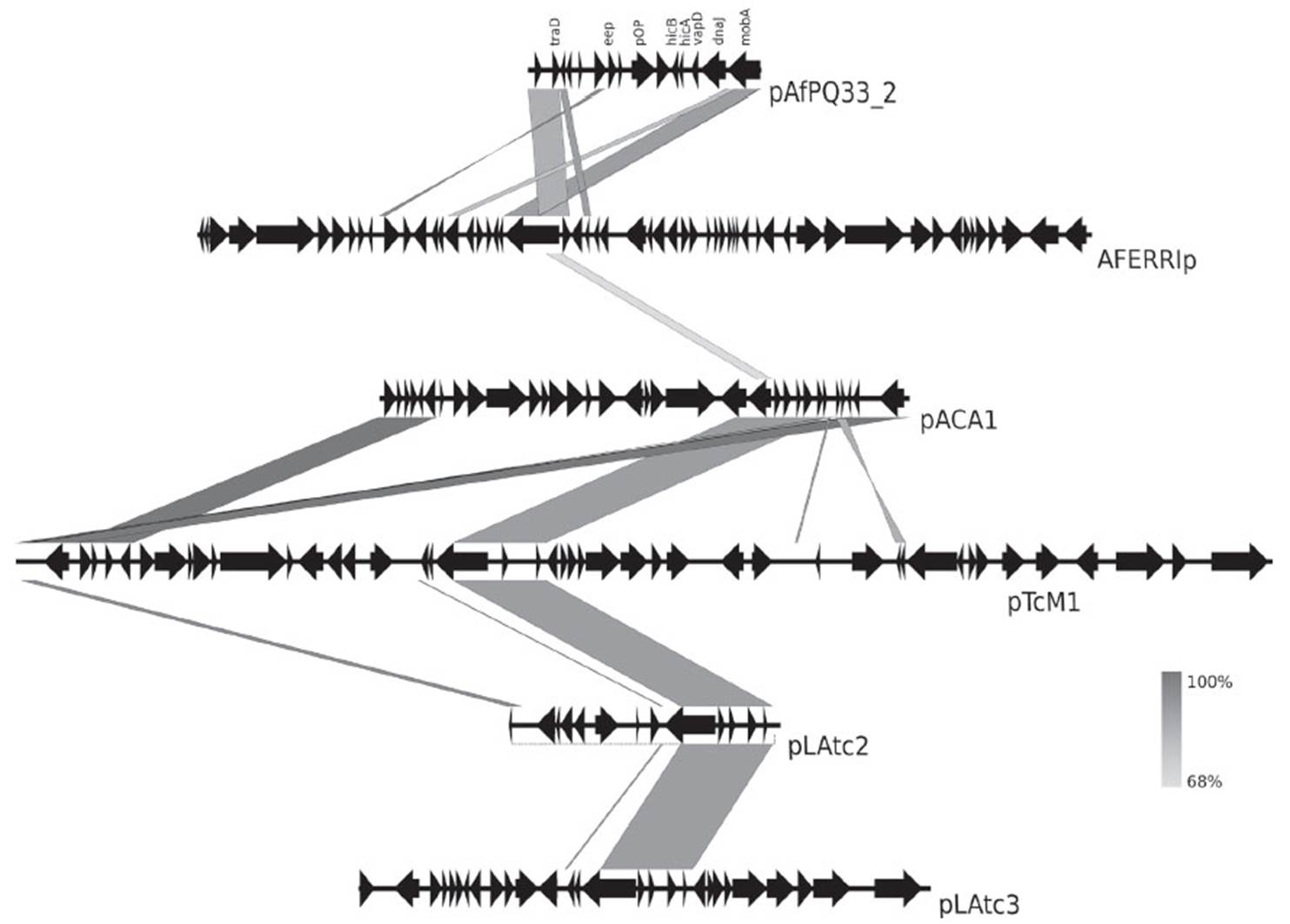
The comparative genomic analysis was carried out using pTC-F14, pTcM1 plasmids, megaplasmid such as mpAca1.1, pACA1.1, pACA1.2, pLAtc1, pLAtc2 and pLAtc3 from A. caldus; pTF4 and pTF5 from A. ferrooxidans; and CF27 from A. ferrivorans. The program EasyFig was used to visualize the organization and comparison of Acidithiobacillus plasmids (Sullivan et al. 2011).
Results and discussion
General characterization
The size of pAfPQ33-1 and pAfPQ33-2 plasmids were 10218 bp and 12094 bp, respectively (Figure 1). The general features of both plasmids are shown in Table 1. The detailed characteristics of each ORF are summarized in Table 2 and Table 3. pAfPQ33-1 plasmid contained 10 predicted ORFs (Figure 2), seven of which (70%) had a function assigned, while three (30%) were conserved hypothetical proteins. With regard to the second pAfPQ33-2 plasmid (Figure 3), 15 ORFs were identified as putative proteins. From the genes predicted, eight (53%) were assigned a putative function, three (20%) were matched to some conserved hypothetical protein, and four (27%) were hypothetical proteins with no database match. The GC skew of the plasmids is shown in Figures 2 and 3. The change of GC skew indicated the plasmids’ origin of replication.
Proteins involved in conjugation
To characterize the plasmid genomes, we describe the probable function of both identified and putative proteins. Among the genes encoding putative proteins involved in conjugation, we found the putative conjugation-related TraD protein in both plasmids, with more than 40% identity to the homologous proteins in A. ferrooxidans to pAfPQ33-1 plasmid. Protein TraD belongs to the type IV secretion system in the inner membrane of gram-negative bacteria and is directly involved in DNA transfer in F+ bacteria (Panicker and Minkley, 1992).
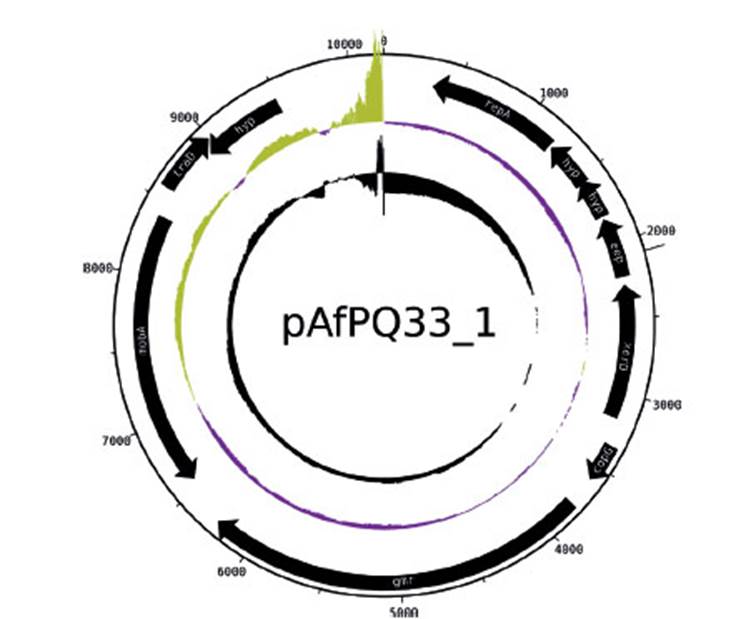
Figure 2 pAfPQ33-1 plasmid from A. ferrivorans represented as a circular plasmid. The first ring (from outside to inside) shows the plasmid size. The internal black arrows indicate its 10 CDS. The second ring indicates the plasmid GC skew, where the origin of replication is indicated (abrupt change of GC skew). The third ring indicates GC content.
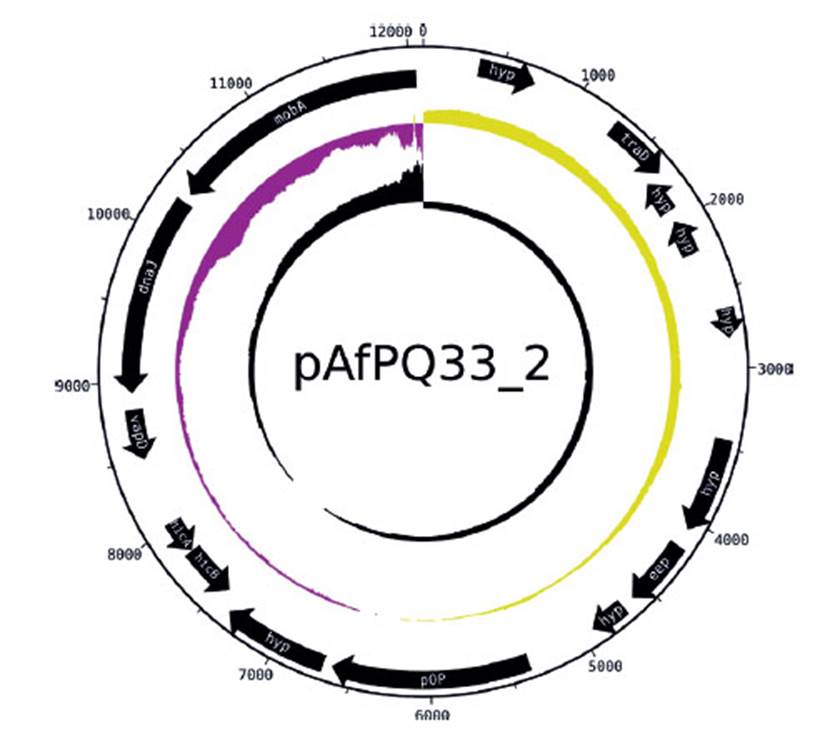
Figure 3 Plasmid pAfPQ33-2 from A. ferrivorans represented as a circular plasmid. The first ring (from outside to inside) shows the plasmid size. The internal black arrows indicate its 15 CDS. The second ring indicates the plasmid GC skew, where the origin of replication is indicated (abrupt change of GC skew). The third ring shows GC content.
Moreover, plasmid pAfPQ33-2 contained the coding sequence for the entry exclusion protein (eep), which plays an essential role as a barrier to DNA transfer among bacteria sharing the same or a closely related F+ factor (Garcillán-Barcia and de la Cruz 2008). Additionally, we found putative mobA homologous genes in both plasmids with more than 40% of identity. This protein was found to be involved in plasmid mobilization in Acidithiobacillus ferrooxidans (Rohrer and Rawlings 1992). While the presence of conjugative plasmids in Acidithiobacillus has been poorly described, the acquisition of mob and oriT sequences has been reported continually by mostly non-conjugative plasmids, such as our plasmids. This fact suggests that conjugative mobilization in Acidithiobacillus is frequent enough, so its plasmids have evolved to take advantage of it, making them potentially mobilizable. This was recently reported in S. aureus (Ramsay et al. 2016). Additionally, we predicted the presence of the tyrosine recombinase XerD in the pAfPQ33-1 plasmid with more than 70% identity to other tyrosine recombinases. This protein could be involved in disentangling chromosomal dimers after bacterial replication, or it could be acting to resolve multimeric forms generated from homologous recombination in a variety of multicopy plasmids (Hallet et al. 1999).
Toxin-antitoxin system
We found a putative hicAB cassette comprising the hicA and hicB genes located in opposing strands in the pAfPQ33-2 plasmid. The hicAB cassette comprises a toxin-antitoxin system that constitutes a translation-independent RNA interference-like system in bacteria and archaea (Jørgensen et al. 2009). In this cassette, at least one hicB gene is always present, and the hicA gene is mostly located adjacent to the hicB gene in the same coding strand; however, sometimes it is positioned in the hicB upstream region in the non-coding strand (Makarova et al. 2006), as observed in plasmid pAfPQ33-2 (Fig. 3). Previous studies of the hicAB locus distribution in numerous bacterial genomes showed that hicAB is transferred horizontally (Makarova et al. 2006). Consistent with this transmission mode, these hicAB cassettes in PQ33 and Sulfobacillus thermotolerans were found to be encoded in plasmids that could serve as the vehicles for horizontal gene transfer (Deane and Rawlings 2011; Li et al. 2016). As previously reported by Makarova et al. (Makarova et al. 2009), HicB contains a helix-turn-helix DNA binding domain and a derivative RNase H fold, while HicA contains a double-stranded RNA binding domain.
Replication origin
The RepA helicase protein was found in plasmid pAfPQ33-1. It shares a high percentage identity (98%) with RepA from Acidithiobacillus thiooxidans plasmid (Table 2). On the other hand, although no replication genes , such as repA, were found in pAfPQ33-2 plasmid (Fig. 3), a putative DnaJ chaperone protein is reported, which has been not only associated with the function of protein re-folding in heat shock or stress conditions (Xiao et al. 2009). DnaJ chaperone protein has been demonstrated to play a role in the initiation of replication with the RepA plasmid protein, thereby forming a dimeric complex that allows the binding of RepA to the origin of replication (Wickner et al. 1991).
In plasmid pAfPQ33-1, the sequence was aligned with pACA1 and pACA2 plasmids from A. caldus ATCC 51756 (Figure 4). A homologous region (86%) of 456 bp that contained the replication origin of the plasmid was found. Additionally, an identity of 85% (175 bp) with pTF91 plasmid from A. ferrooxidans was found. This region is located close to the gene repA. For pAfPQ33-2 plasmid, no successful match with Acidithiobacillus plasmids was noted. However, a GC skew analysis helped identify the replication origin close to the gene mobA in pAfPQ33-2 plasmid, approximately 100 bp downstream to the coding region (Fig. 3). Following the report by Lopez et al. (1999), bacterial and archaeal extremophiles must show these GC skew patterns.
Evolutionary relationships among Acidithiobacillus plasmids
As evidenced in Figure 4, there is an evolutionary relationship among plasmids pACA1 and pACA2 from A. caldus ATCC 51756 and pAfPQ33-1 plasmid. The particular identity of the diguanylate cyclase-phosphodiesterase (DGC-PDE) protein between the plasmids suggests a similar plasmid maintenance mechanism, either horizontal transfer or genome reorganization in bacteria belonging to the Acidithiobacillus genus (Fig. 4). Furthermore, the identity of genes eep, xerD and the two hypothetical genes with those in pACA2 plasmid suggests a common mechanism of genome reordering and plasticity between A. caldus and A. ferrivorans. Moreover, the similarity found among the plasmids of A. caldus M1, pTcM1, and those from the other strain, pACA1, pLAtc2 and pLAtc3, with pAfPQ33-2 plasmid, reinforce the hypothesis of shared evolutionary traits (Figure 5). Liljeqvist et al. (2015) reported the metagenome of Acidithiobacillus from extremely cold environments and found an integrative community that could support the shared evolutionary traits among species. The exchanging of plasmids between species might confer this function.
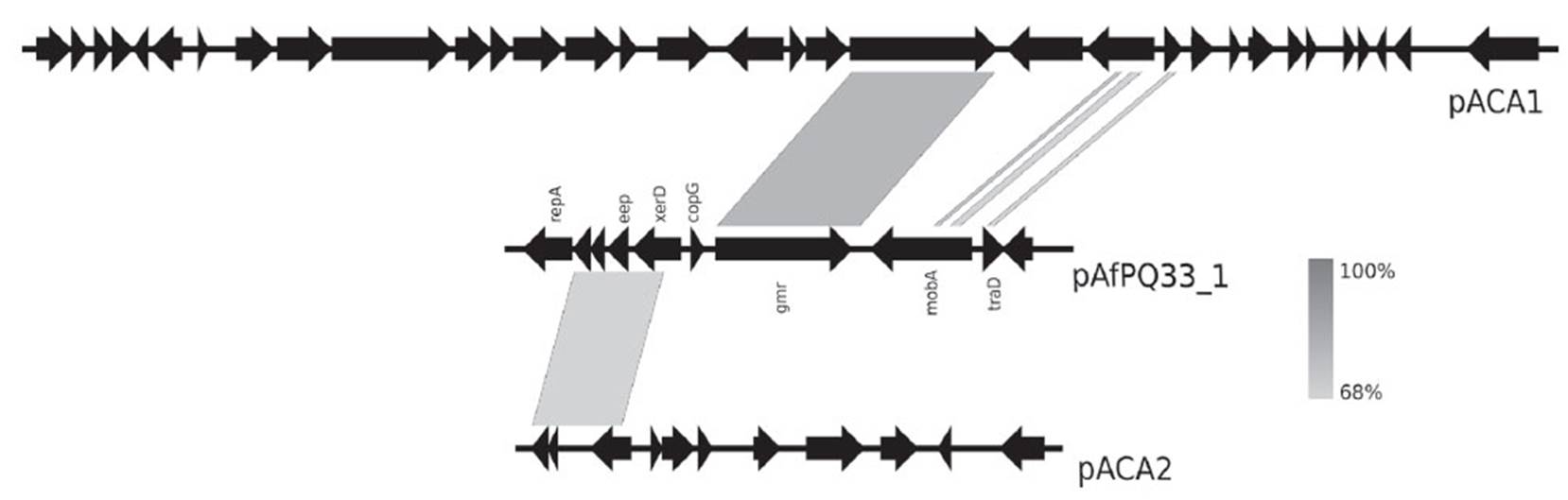
Figure 4 Comparative analysis of pAfPQ33-1 plasmid from A. ferrivorans PQ33 and pACA1 and pACA2 plasmids of A. caldus ATCC 51756.
Proteins involved in environmental adaptation
Putative endoribonuclease VapD
The coding sequence for the VapD protein (belonging to the CAS2 protein family) was found in plasmid pAfPQ33-2. It presented an identity of 65% compared with endoribonuclease from Pseudomonas sp. This protein acts as a protective element against extracellular genetic element invasion (Beloglazova et al. 2008), and it is likely to have an important role in the PQ33 strain because horizontal transfer capacity has become an advantage for Pseudomonas species. Since many proteins from the family CAS2 have been related to mRNA degradation, it is suggested that vapD could play a role as an endoribonuclease that is mRNA specific (Beloglazova et al. 2008).
Putative porins O and P and phosphate-selective proteins
We identified the phosphate-selective porins O and P proteins in pAfPQ33-2 plasmid. These present an identity of 68% (417 aa) compared with the porin protein present in the chromosome of A. ferrivorans, which coincides with other phosphate porins found in the Acidithiobacillus genus. The predicted domain organization of porins O and P is shown in Figure 6. During bioleaching processes, phosphate starvation generates activation of genes encoding proteins that allow the recovery of environmental phosphate (Bobadilla Fazzini and Parada Valdecantos 2009), including selective phosphate porins O and P, which have a specific phosphate/orthophosphate binding site and are overexpressed under conditions of starvation (Hancock et al. 1982; Poole et al. 1987). Thus, the specific location of the gene of this protein in the plasmid could be involved in the adaptation of A. ferrivorans to the environmental stress conditions. This suggests rapid adaptation of this specie, as evidenced in their ability to grow in vitro compared to A. ferrooxidans.
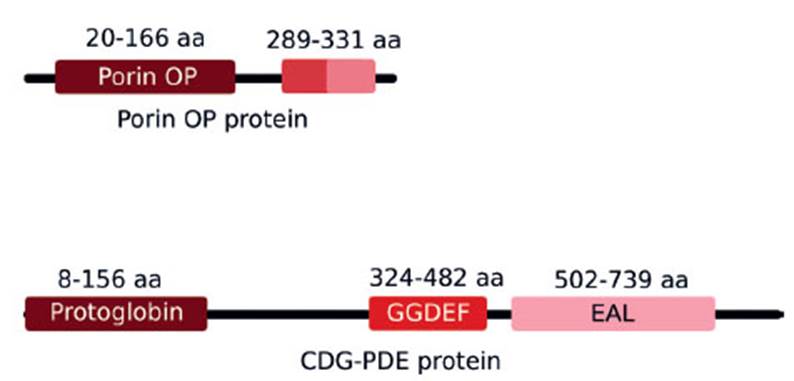
Figure 6 Domain organization of porins O and P and CDGPDE proteins. a) Putative porins O and P aligned to positions 20-166 (E-value = 5.75e-05) and 289-331 (E-value = 4.6e-05) of a 359 residue-long Hidden Markov Model (HMM) defining a phosphate-selective porin O and P family. b) CDG-PDE aligned with high confidence to three HMMs defining a protoglobin family (Evalue = 2.8e-47), and GDEF (Evalue = 6.7e-21) and EAL domains (Evalue = 6.3e-67)
Diguanylate cyclase-phosphodiesterase (DGCPDE) with a chemoreceptor zinc-binding domain
Recently, diguanylate cyclase-phosphodiesterase (DGC-PDE) has been studied due to its role in adherence during the biomining process. In A. caldus and A. ferrooxidans, the protein has been expressed in bacteria growing over sulfide surfaces, and the efficiency of bioleaching was noted. Additionally, in A. caldus mutant of this protein, no evidence of adherence to sulfur surfaces was noted (Castro et al. 2015). Our plasmid pAfPQ33-1 has an ORF encoding the DGC-PDE protein that can hydrolyse and synthetize c-di-GMP with the presence of both GGDEF and EAL domains. It has an identity of 78% with GGDEF/EAL domains of A. ferrivorans SS3. However, we identified GGDEF, EAL and protoglobin domains in Pfam and Swiss-Model (Fig. 6). Surprisingly, a chemoreceptor zinc binding (CZB) domain adjacent to the GGDEF-EAL domains was predicted; however, it was found in a different open reading frame and is similar (63% of identity) to that present in A. caldus. The A. ferrivorans draft genome reports approximately 16 putative coding-genes for the enzyme DGC-PDE; many of them present GGDEF and EAL domains, as well as a series of sensor domains, and a CZB domain. Studies in E. coli have shown that biofilm formation was dependent on the DGC-CZB activity, which was modulated by the availability of zinc, since mutation of amino acids residues that coordinate this ion generated a decrease in exopolysaccharide formation (Zähringer et al. 2013).
Conclusions
In a genomic context, the analysis of plasmids from PQ33 strain suggests their key role in the horizontal transfer system of A. ferrivorans. The comparison between plasmids of A. ferrivorans and A. caldus shows that there are evolutionarily shared traits involving the gene exchange mechanism among species. These findings could be significant for a global understanding of the environmental role of the species. Additionally, the characterization of the plasmids offers new insights into the development of vectors for the psychrotolerant strains. The description of the metabolic diversity in psychrotolerant Acidithiobacillus spp. is becoming vital for the understanding of bioleaching operations in cold climate zones such as the South American Andes











 uBio
uBio