INTRODUCTION
COVID-19 is a serious global public health problem, with more than 657 million confirmed cases and more than 6.68 million deaths worldwide as of 25 December 2022 (1). The rapid spread of the disease represents a major challenge for developing countries that must contend with economic and logistical gaps for timely diagnosis of cases 1).
The gold standard test for the diagnosis of SARS-CoV-2, according to the World Health Organization (WHO), is the real-time reverse transcription-polymerase chain reaction (RT-qPCR) test, which requires high-level infrastructure, sophisticated equipment, high-cost reagents and, despite technological advances, the time required for the entire process is extensive (~4 to 6 hours), which makes this technique difficult to implement in the different regions of the country 2).
In view of this problem, there is an urgent need to use alternative cost-effective techniques to aid in the diagnosis of COVID-19, such as the reverse transcription loop-mediated isothermal amplification (RT-LAMP) 3,4 and cross-priming amplification (CPA) methods 5). Currently, both methods are used in Callao for the mass screening of SARS-CoV-2; nonetheless, it is uncertain if they are equivalent techniques, possibly leading to misguided results.
The RT-LAMP technique involves the use of 4 to more primers; and can provide results in short periods of time. This technique is more feasible to implement, as it uses simpler equipment and infrastructure (6, 7) and was validated for Peru in 2020 giving a sensitivity and specificity of 87.4% and 98.8%, respectively (8). The CPA performs the process in less time (~45 minutes vs 60-85 minutes of RT-LAMP), high levels of sensitivity and specificity and multi-modules streamlining the processes 9). Both techniques allow timely detection of SARS-CoV-2 and are available in Peru; nonetheless, CPA feasibility was not previously evaluated.
The aim of this research study is to compare the RT-LAMP and CPA methodologies with respect to the gold standard technique, RT-qPCR, in order to determine if both techniques are equivalent for SARS-CoV-2 screening.
MATERIALS AND METHODS Study design
Paired comparative case-control design, where nasopharyngeal swab samples stored at -20°C in the Regional Laboratory of Molecular Biology of DIRESA-Callao (LRBM) previously tested for SARS-CoV-2 by RT-qPCR from June 2021 - December 2021 were selected. The LRBM as a public laboratory daily receives nasopharyngeal samples that comes from SARS-CoV-2 diagnosis or screening requests.
The sample selection was done using a de-identified list given by the LRBM; this list contained a code and its result of SARS-CoV-2 detection. Afterwards, the samples were chose under systematic random sampling. The selected samples were aliquoted in two sterile cryotubes. One aliquot was used for RT-LAMP determination and the other one for CPA assay. The index test is the CPA, and the comparator test was the RT-LAMP.
Cases were defined as patients who tested positive for SARS-CoV-2 by real-time RT-PCR. Controls were defined as patients who tested negative for SARS-CoV-2 by real-time RT-PCR.
The following patients/samples were excluded: samples of patients <18 years-old, patient whose sample was indeterminate for SARS-CoV-2 by real-time RT-PCR, patient whose sample did not show signs of good storage and patient whose sample contained less than 1 mL. A total of 21 were excluded because of the age criteria. A total of 200 samples were considered for the study, of which 100 were used for both LAMP and CPA analysis, and 100 additional samples for the CPA analysis (200 in total).
Extraction of nucleic acids (RNA)
RNA extraction was carried out using the QIAamp Viral RNA Mini Kit 250 extraction kit (QIAGEN) for the RT-LAMP method. It consists of optimized enzymes and buffers that lyse samples, stabilize nucleic acids and enhance selective RNA uptake on the QIAamp membrane. To ensure RNA integrity, samples are lysed under highly denaturing conditions to inactivate RNAases. Alcohol is added, and the lysates are loaded onto the QIAamp spin column. Wash buffers are used to remove impurities and then pure, ready-to-use RNA is eluted in water or low-salt buffer, all according to the manufacturer's instructions. RNA simples were stored at -20°C until use. During extraction, the simples were coded, a nomenclature that was maintained throughout the labelling process, including elution. Coding was carried out by a principal investigator, who listed the selected samples with the corresponding new code. This will prevent traceability by the analyst, and will therefore be blind to the RT-qPCR results 10).
Detection of SARS-CoV-2 by RT-qPCR
Cases and controls were identified based on the RT-qPCR assays performed in the Regional Laboratory of Molecular Biology of DIRESA-Callao. In brief, analyses were performed with the SARS-CoV-2 PCR Kit (Lifotronic, China) using primers and probes for SARS-CoV-2 detection and an internal control (RNAsa P) (green channel, FAM: 470-510 nm and orange, VIC: 585-610 nm, respectively). They were considered positive when cycle threshold (Ct) values < 37 (FAM) and Ct < 40 (VIC) were obtained, respectively; this was in accordance with the manufacturer's indications 11).
Detection of SARS-CoV-2 by RT-LAMP RT-LAMP reactions were performed according to Lamb et al. (12), using WarmStart® Colorimetric LAMP 2X Master Mix DNA and RNA (Eiken Chemical Co., Ltd., Tokyo, Japan), which contains a pH indicator that allows for colorimetric visualization. 44 µM of the primers FIP (16 µM), BIP (16 µM), F3 (2 µM), B3 (2 µM),
LOOP F (4 µM), BUCLE B (4 µM), and 56 µM of water will be used; in addition, 20 µL of the reagents MIX-LAMP (12.5 µL), MIX- Primers (2.5 µL), RNA (5 µL), and 5 µL of water will be used. The amplification reaction was carried out at 65 °C for 45 minutes, followed by an inactivation phase of the reaction at 80 °C for 5 minutes. RT-LAMP processes uses 20ul of LAMP reagent and 5ul of sample, with 25ul being the final volume 8).
A negative result is evidenced by the absence of red to yellow color change; a positive result is evidenced by the solution turning yellow. The results shall be read by the same analyst who performed the RNA sample extraction.
Detection of SARS-CoV-2 by CPA
CPA reactions were performed on the USTAR® brand EasyNAT™ System equipment, which is based on three-stage magnetic conductivity extraction technology and patented isothermal cross-priming amplification technology.
This technology makes use of a two-stage reaction tube, the first being a column where the genetic material is extracted by adding 500 µL of sample with 500 µL of magnetic beads. The second phase is the elution medium where the reading is done by means of a fluorescent signal.
The reaction column is entered into the equipment, being identified according to the code provided by the researcher. Once inside, isothermal amplification is carried out, with constant quantification of a fluorescent signal emitted by the médium if viral RNA is present. Once the process is finished after 40 minutes, the same interface will give a positive or negative result, depending on the case 9).
Statistical análisis
The STATA 17.0 package was used for statistical analysis. True positives and true negatives were determined by comparing the results obtained by RT-LAMP or CPA with the gold-standard reference test (RT-qPCR). This was used to calculate the sensitivity and specificity of the index test and the comparator test.
As the samples tested are paired or related, McNemar's test was used to compare the sensitivity and specificity of both tests. Subsequently, the observed concordance percentage was calculated; as well as the kappa statistic to determine whether the observed concordances are higher than those expected purely by chance. Kappa bias corrected was calculated by bootstrapping set to 1000 repeats. A p<0.05 will be considered statistically significant.
RESULTS
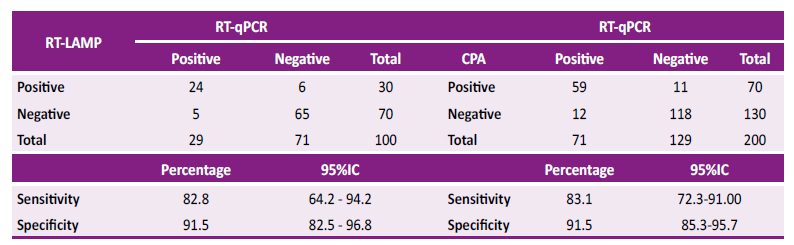
For the RT-LAMP method 29 positives (cases) and 71 negatives (controls) were obtained. RT-LAMP was able to identified 30 positives and 70 negatives for SARS-CoV-2. When evaluating the sensitivity and specificity of the RT-LAMP technique with reference to the gold-standard, a sensitivity of 82.8% (95%CI 64.2 - 94.2%) and a specificity of 91.5% (95%CI 82.5 - 96.8%) were identified, as shown in Table 1.
For the CPA method, a total of 71 positives and 129 negatives were used. It identified a total of 70 positives and 130 negatives. When compared with the results obtained by RT-qPCR, a sensitivity of 83.1% (95%CI 72.3 - 91.0%) and a specificity of 91.5% (95%CI 85.3 - 95.7%) were obtained, as shown inTable 2.
Table 2. Agreement and kappa index between CPA and RT-LAMP
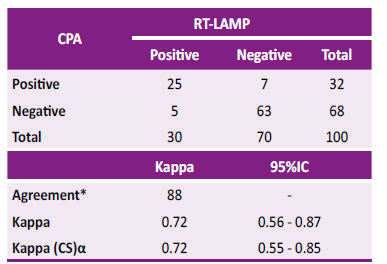
* Percentage agreement between CPA and RT-LAMP tests is shown.
α Kappa (CS): Kappa corrected for bias by bootstrapping at 1000 replicates
When comparing both tests (RT-LAMP and CPA) with a total of 100 samples, a concordance of 88.0% and a Kappa index of 0.72 (0.56 - 0.87) were obtained; when correcting the Kappa index for bias using the resampling technique by bootstrapping set to 1000 replicates, a 95%CI of 0.55 - 0.85 was obtained (Table 2). In the comparison of sensitivity and specificity between CPA and LAMP, McNemar’s chi-squared test for paired sample showed no statistical difference -0.02 (95%CI -0.09, 0.05, p=0.563).
DISCUSSION
Callao reported the highest mortality rates at the national level, 10 deaths per 1,000 inhabitants 12). In the Callao region there are different international and national access points such as sea, air and land ports, which is why epidemiological monitoring based on short-term molecular screening is crucial to control the disease. Therefore, two isothermal molecular screening methods were evaluated: RT-LAMP and CPA compared to the gold standard, RT-qPCR. Each of them has different utilities and methodology.
The importance of applying molecular screens is that they can detect the presence of virus or its remnants 5 at any stage of the disease, because these screens have been shown to detect high, medium and low viral loads, unlike serological tests because performance is influenced by symptomatology, days of infection, or virus lineage and they are not confirmatory tests for the disease 13-15).
The evaluated results show that both techniques have adequate and equivalent values of sensitivity (RT-LAMP: 82.8%, CPA: 83%) and specificity (RT-LAMP and CPA: 91.5%), as well as a high concordance (88%) between both techniques. Our results are in line with previous studies such as that of Lu et al. 5 showed that the CPA technique shows high sensitivity and specificity versus the gold standard, been able to detect from 100 copies of ARN per reaction tube. On the other hand, Rai et al. 16 based on a systematic review of different molecular techniques for SARS-CoV-2 detection found that RT-LAMP is capable to detect from 4 copies/uL.
The CPA method, based on isothermal amplification reactions mediated by a DNA polymerase, avoids additional steps such as denaturation or additions of other enzymes 17). On the other hand, the LAMP reaction is also an isothermal methodology, characterized by the pH change generated by the chelation of Mg2+ ions from dNTPs and modification of pH indicators such as phenol red and hydroxynaphthol blue, providing results in less than 1 hour 3).
Although the most widely used technique after RT-qPCR is RT-LAMP, this is due to its ease of implementation in remote and rugged locations, because the equipment for this technique is minimal and inexpensive, as opposed to the CPA technique. The implementation of the CPA test is a highly viable alternative due to its high sensitivity and specificity, it is a bit complex due to the lack of supplies and expensive equipment, but the benefits in the medium and long term make it an equivalent alternative applicable to areas of difficult access where it is easy to perform an RT-qPCR test. Since it is necessary to have laboratory tests with an adequate diagnostic performance that is comparable with the RT-qPCR test, thus closing the gaps of the existing demands 8).
A limitation of the study was that we were only able to use 100 samples for the LAMP analysis of sensitivity and specificity and, therefore, for the kappa assessment; however, the bootstrapping procedure allowed us to reduce this error in the kappa estimation.
In conclusion, it is evident that RT-LAMP and CPA tests are equivalent and suitable for the screening of SARS-CoV-2, thus demonstrating the great potential that they have, as well as the possibility of expanding the range of clinical tests.