Introduction
The genus Staphylococcus currently comprises more than 80 species and subspecies according to the website of the List of Prokaryotic names with Standing in Nomenclature by Leibniz Institute DSMZ-German Collection of Microorganisms and Cell Cultures GmbH (https://lpsn.dsmz.de/genus/staphylococcus, consulted on March of 2020). Bacteria belonging to this genus are generally Gram-positive, nonmotile, aerobes, catalase-positive, oxidase-negative and able to grow between 18 and 40 °C (Oh et al., 2019; Schleifer and Bell, 2015). Regarding the phylogeny, the genus Staphylococcus belongs to the Staphylococcaceae family and the Bacillales order, which shares with species of different genera such as Bacillus, Listeria, Paenibacillus as well as Macrococcus, Enterococcus, Streptococcus, and Lactobacillus all as part of the phylum Firmicutes (De Vos, 2015; Schleifer and Bell, 2015).
Besides the importance of Staphylococcus as a pathogenic agent of humans and several animals species, from fish to cattle, they could also act as infective agents of plants under certain conditions as described in Arabidopsis thaliana (Canovas et al., 2016; Oh et al., 2019; Prithiviraj et al., 2005). Moreover, apart from the conventional sources of staphylococci mainly recovered from clinical samples extracted from human and animal skin, glands, and mucous membranes, staphylococci can be isolated from diverse sources such as air, water and soil (Correia et al., 2019; Kozajda et al., 2019; Mandal et al., 2015; Schleifer and Bell, 2015).
The importance of staphylococci go beyond their role as pathogens due to their capacity to produce antimicrobial peptides (Bastos et al., 2009), their role in meat fermentation processes (Stavropoulou et al., 2018) and mainly because of the beneficial effects on plants of some strains that inhabit the rhizosphere, as described for mangrove trees and Leptochloa fusca (Holguin, 1992; Shahid et al., 2019). These strains could be considered as rhizobacteria owning attributes related to plant growth promotion such as the capacity to fix nitrogen as well as to produce phytohormones and specific compounds (Holguin, 1992; Shahid et al., 2019).
Capsicum annum L. is an important crop in Peru. Members of the genus Capsicum belong to the Solanaceae family and are native of subtropical and tropical areas in America (Tripodi and Kumar, 2019). The cultivar Piquillo is one of five domesticated crops of C. annuum which is widely cultivated and studied along with its two variants: hot pepper as a spice crop and sweet pepper as a vegetable crop (Jindal et al., 2020; Khoury et al., 2020). C. annuum cv. Piquillo is an important export crop in Peru which sends this commodity as raw material and other derivate products to the European and American market (Agraria.pe, 2019; MINAGRI and ProInversion, n.d.). In recent years, along with the promulgation of the Plan of sustainable development of species of the Capsicum genus 2018-2028: peppers and chilies, devised by The Ministry of Agriculture of Peru, greater interest has been taken into this crop (MINAGRI, 2017). Therefore, the objective of this research was to identify Staphylococcus isolates from rhizospheric soil samples of C. annuum cv. Piquillo.
Materials and methods
Bacteria were isolated from rhizospheric soil samples of cultivated Capsicum annuum L. cv Piquillo areas under open-field conditions in January 2018 in Viru, La Libertad, Peru (08°24’18” S, 78°51’18” W). Samples were collected and kept under refrigeration during transportation to the laboratory and stored at 4 °C. Then, morphologically distinct bacteria were isolated by serial dilutions and cultured in Tryptic soy agar (TSA) and Reasoner’s 2A (R2A) agar plates at 28 °C for up to five days. Isolates were labeled as Ca2, Ca5, Ca6 and Ca7. After, bacterial genomic DNA extraction from each pure isolate, 16s rRNA gene amplification, sequencing and bioinformatic analysis were performed according to the protocol described by Belgini et al., 2014. Briefly, genomic DNA was extracted following the CTAB method and DNA integrity and concentration were estimated through electrophoresis in 0.8 % agarose gel stained with SYBR Safe 10.000x in DMSO (Invitrogen). DNA obtained was used in polymerase chain reaction (PCR) reactions for amplification of 16S rRNA using the primers 10f (5'-GAGTTTGATTCAGGCCCTG-3') and 1100r (5'-GTTGTGAGGGTTGGGG-3'). PCR products were purified using mini-columns (GFX PCR DNA and Gel Band Purification Kit, GE Healthcare) and subjected to sequencing in an automated sequencer ABI 3500XL Applied Biosystems™. Partial 16S rRNA consensus sequences were obtained using the BIOEDIT software (Hall, 1999) and were compared with those of reference type strains available in EZBioCloud (https://www.ezbiocloud.net/) and RDP (Ribosomal Database Project, Wisconsin, USA https://rdp.cme.msu.edu/) databases. The sequences were aligned using the CLUSTAL X program (Thompson et al., 1997) and analyzed with MEGA 7 software (Kumar et al., 2016). The evolutionary distances were calculated using the Kimura DNA substitution model (Kimura, 1980) and the phylogenetic reconstruction was done using the neighbor joining (NJ) algorithm (Saitou and Nei, 1987), with bootstrap values calculated from 1,000 replicates.
Results and discussion
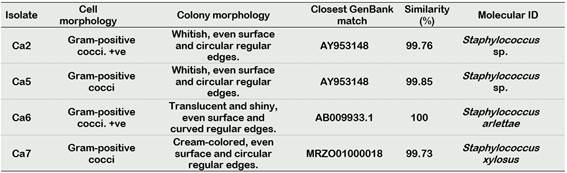
We identified four bacterial isolates from the genus Staphylococcus not previously reported in C. annuum rhizospheric soils: Isolate Ca2 and Ca5 which both match to Staphylococcus sp., isolate Ca6 to Staphylococcus arlettae and isolate Ca7 to Staphylococcus xylosus (Table 1).
The presence of Staphylococcus in rhizospheric soils could be explained as bacteria from that genus have been described as ubiquitous, being isolated from many sources such as beach sand, sea water, fresh water, plant surfaces and products, feeds, meat and poultry, dairy products, dust, air and soil (Schleifer and Bell, 2015). Nonetheless, studies reporting Staphylococcus present in rhizospheric soils are still scarce. López et al. (2009) found a mangrove rhizosphere-associated Staphylococcus sp., Leiva et al. (2013) reported a phosphate-solubilizing S. vitulinus present in the rhizosphere of cacao, and Shahid et al. (2019) identified two Staphylococcus strains which promoted the growth of maize under salt stress.
Table 1 Morphological description and molecular identification of Staphylococcus isolates from rhizospheric soil samples of C. annuum cv Piquillo. Cell and colony morphology of each isolate are described. Closest GenBank (www.ncbi.nlm.nih.gov/) match’s accession number and similarity percentage are shown. +ve. Gram-variable
Regarding S. arlettae and S.xylosus, the former has been isolated from poultry and goats, but has also been reported in cardamom rhizosphere as a bacteria with bioremediation potential to degrade fipronil residues (At et al., 2019; Schleifer and Bell, 2015); the latter, is able to grow in habitats that contain only an inorganic nitrogen source and thus could be more free-living than other staphylococci; besides, it has been isolated from beach sand, natural waters, marsh grass, and plant products, and has also been found in the rhizosphere of potato (Berg et al., 2005; Schleifer and Bell, 2015).
Studies concerning rhizospheric microbiome profiling, isolation and identification of rhizobacteria from C. annuum report bacteria mainly from Gammaproteobacteria and Bacilli classes, Serratia and Bacillus genera (Asaff-torres et al., 2017; González et al., 2017). Particularly, S. aureus has been reported in the rhizosphere of cultivated C. chinense (Chinakwe et al., 2019). Despite the above, we have not found reports of Staphylococcus present in rhizospheric soils of C. annuum. The isolates found in the present research could have the potential to promote plant growth or to be utilized for ecological, industrial and agricultural purposes, therefore, further studies to assess their role in the rhizosphere and their effect on Capsicum and other crops are suggested.
Conclusions
Not previously reported Staphylococcus sp., Staphylococcus arlettae and Staphy-lococcus xylosus isolates from C. annuum rhizospheric soils were identified. These isolates could have potential applications in agriculture. Further studies to determine their ecological role in the rhizosphere as well as their potential applications in antimicrobial development focusing on antimicrobial peptide production, food processing regarding meat fermentation processes and pesticide biodegradation as reported for fipronil residues in soils are recommended.