1. Introduction
Peru is the eighth largest coffee producer in the world (16) and the United States is the main export destination (7; Nolte, 2020). Peruvian coffee is produced in 10 regions between 600 and 1,800 meters above sea level (m.a.s.l.), with a total crop extension of 375,000 ha (Junta Nacional del Café, 2019).
Nowadays, Peruvian coffee production is recovering from coffee leaf rust (CLR) caused by Hemileia vastatrix (Hv), which affected 50 percent of the 2013/14 harvest (Nolte 2020; McCook and Vandermeer 2015). Faced with the Peruvian coffee crisis, National Institute of Agricultural Innovation (INIA) established a Coffee Germplasm Collection (INIA-CGC) to provide valuable information for its genetic conservation and breeding programs. INIA-CGC located at Pichanaki Agricultural Experimental Station in Junin, it maintains 169 coffee accessions collected from the main coffee growing regions: Amazonas, Cajamarca, Huanuco, Junin, Pasco and Ucayali.
Hv is a mandatory biotrophic fungus that specifically attacks coffee plants (Talhinhas et al., 2017) and causes premature defoliation in several species of the genus Coffea, of which C. arabica is the most susceptible (Silva et al., 2018). The success in the infection of Hv is due to its large genetic diversity that manages to overcome the resistance of its host. The different races of Hv are classified according to their ability to infect different coffee materials that have different combinations of resistance genes (Talhinhas et al., 2017). Currently, the Centro de Investigação das Ferrugens do Cafeeiro (CIFC) of the Instituto Superior de Agronomia (ISA) - Universidade de Lisboa (U. Lisboa) has around 3,600 samples of Hv from 40 coffee-producing countries, which are classified in more than 50 breeds of Hv for a selected range of 23 differential coffee plants (12). The CIFC breed nomenclature is attributed to isolates with distinct and unique combinations of nine virulence genes as inferred in gene-by-gene theory and are described as sequential Roman numerals in order of their detection (Talhinhas et al., 2017). For example: race I has a virulence gene (V2,5); race II (V5); race III (V1,5); race X (V1,4,5); race XIII (V5,7); race XVII (V1,2,5); race XXII (V5,6); race XXIV (V2,4,5); race XXVIII (V2,4,5,6); race XXXVI (V2,4,5,8); race XXXVII (V2,5,6,7,9) and race XXXIX (V2,4,5,6,7,8,9). Some of the races mentioned before were reported in Brazil (Zambolim, 2016); Costa Rica and Guatemala (Avelino et al., 2015).
Up to 9 dominant resistance genes have been identified in the Coffea genus (SH1 to SH9) and correspond to the nine Hv virulence genes (Mahé et al., 2008). Of these resistance genes, SH1, SH2, SH4 and SH5 have been found in C. arabica, but they have not provided lasting resistance to most Hv races. The other resistance genes, SH6, SH7, SH8 and SH9, found in C. canephora, have been introgressed into C. arabica, to obtain resistant hybrids (Mahé et al., 2008). Finally, the SH3 gene found in C. liberica (Prakash et al., 2004) seems to provide lasting resistance to the coffee plants that have it.
Following the discovery of a rust-resistant coffee plant on the island of Timor (Timor Hybrid, a spontaneous interspecific hybrid between C. arabica and C. canephora), coffee improvement programs were developed using this hybrid as the main gene source to obtain varieties resistant to coffee rust (Talhinhas et al., 2017; Romero et al., 2014). Despite of their early success which allowed the release of highly resistant and productive cultivars, most of them became susceptible due to the appearance of more aggressive strains of Hv (Talhinhas et al., 2017; Cabral et al., 2016).
Another natural hybrid (S.26) between C. arabica and an unknown diploid coffee species was reported in India. This hybrid was used as the main source of resistance to rust in coffee improvement programs in India, from which the hybrid S.795 showed high levels of tolerance in the field for races I and II of Hv. More recently, it has been revealed that coffee plants from India had the SH3 resistance gene for coffee rust from Coffea liberica (Prakash et al., 2002).
Numerous studies have used molecular markers to understand the introgression of C. canephora or C. liberica resistance genes (Ribas et al., 2011; Prakash et al., 2002; Lashermes et al., 2000). These markers allow the rapid and early identification of plants carrying Hv resistance genes through marker-assisted selection (MAS). Moreover, the use of molecular markers accompanied with a high-throughput separation method as capillary electrophoresis (CE) have allowed the develop of a rapid procedure to identify alleles (Durney et al., 2015).
Among the molecular markers associated with resistance to coffee leaf rust, Mahé et al. (2008) reported SCAR, SSR and BAC markers developed from populations that carry the SH3 gene. These markers, used by Alkimim et al. (2017), Valencia et al. (2017) and González-Martínez et al. (2009), are able to discriminate presence and absence of the SH3 gene in different hybrid progenies of C. liberica and C. arabica.
In this work, four markers linked to the SH3 resistance gene (Alkimim et al., 2017; Valencia et al., 2017; González-Martínez et al., 2009; Mahé et al., 2008) were used to screen the accessions from INIA-CGC, and identify potential accessions carrying the SH3 gene in order to implement a coffee breeding program in Peru.
2. Materials and methods
Biological material
Coffee samples from INIA-CGC. The coffee germplasm collection has 169 accessions, from which 102 accessions had variety data (denomination according to producers, Figure 1) and 67 accessions without variety denomination (Amazonas:1; Cajamarca: 5; Huanuco: 2; Junin: 34 and Pasco: 25 accessions).
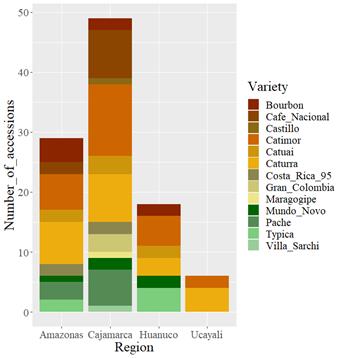
Figure 1 Geographic distribution of coffee varieties from INIA’s Coffee Germplasm Collection. Variety denomination according to producers.
The method used to collect the samples consisted of taking leaves of the superior third of three plants (biological replicates) per accession; except for the accessions CA-164 and CA-165 that have only two biological replicates. A total of 505 samples were collected, stored with silica gel, and transported to INIA’s Laboratory of Molecular Biology and Genomics (Lima, Peru) for further sample processing.
DNA extraction. Genomic DNA was isolated from dehydrated young leaf tissue by two methods: the CTAB method (Doyle & Doyle 1987) and a method provided by Dr. Lashermes from IRD-France (personal communication), for those samples that had poor quality after extraction with the CTAB method. Dr. Lashermes’ protocol used 75 mg of dehydrated leaves that were snap-frozen with liquid nitrogen and ground, then 400 µL of the extraction buffer was added (1.5 M NaCl; 200 mM Tris-HCl, pH 8; 40 mM EDTA, pH 8; 3% sarkosyl; 1% sodium bisulfite; 2% MATAB, the last reagent was added before using the buffer) and 400 µL of the lysis buffer (350 mM sorbitol; 200 mM Tris-HCl, pH 8; 40 mM EDTA, pH 8; 2% PVP, the last reagent was added just before using the buffer) and incubated at 62 °C for 30 min with regular mixing by inversion. After incubation, the mix was cooled to room temperature and 1 ml of chloroform: isoamyl alcohol solution (24:01) was added and mixed then was centrifuged for 5 min at 12,000 rpm. The supernatant was transferred to new 2 ml tubes and 20 µL of RNAse (10 mg/ml) was added, then placed on a heat block at 37 °C for 30 min. After this period, a volume of isopropyl alcohol was added to precipitate the DNA and centrifuged at 12,000 rpm for 5 min.
The supernatant was carefully discarded, and the DNA pellet was washed with 200 µL of ethanol (70%) and centrifuged at 12,000 rpm for 3 min. When finished, the ethanol was discarded, and the pellet could dry. Finally, the pellet was resuspended in 50 µL TE at 4 °C overnight and storage at -20°C. The quality of the DNA obtained from the two mentioned protocols was assessed by means of a 1% agarose gel with SYBR Safe (Invitrogen) in TAE 1X buffer, and it was visualized under UV light. The DNA obtained was quantified by spectrophotometry on an EPOCH (BiotekTM). In addition, DNA from EA67 (C. liberica, var. Liberica) and the hybrid S.288 (C. arabica x C. liberica), kindly provided by Dr. Lashermes, were used as positive controls for molecular markers linked to the SH3 gene.
Marker Amplification and capillary electrophoresis. Four molecular markers reported by Mahé et al. (2008) as linked to the SH3 gene of C. liberica were used to screen 169 accessions of coffee plants using capillary electrophoresis. To do this, the 19 bp M13 tail (5'-CACGACGTTGTAAAACGAC-3') was added to the 5' end of every sense primer (Table 1). PCR conditions were similar to those described by Mahé et al. (2008), with some modifications mentioned by Schuelke (2000). PCR reactions were done in a 10 µL total volume which contained 40 ng of DNA, 1X Buffer Taq 10X (Thermo Fisher Scientific, USA.), 2 mM MgCl2, 100 µM dNTP (Thermo Fisher Scientific, USA), 0.1 µM sense primer, 0.4 µM antisense primer, 0.4 µM fluorophore, 0.5 U of Taq polymerase enzyme (Thermo Fisher Scientific, USA). The fluorophores used were: FAM for the Sat244 marker, VIC for the BA124-12K-f marker, NED for the BA-48-21O-f marker and PET for the SP-M8-SH3 marker. The PCR program included an initial denaturation stage at 95 °C for 3 min, followed by 30 cycles of denaturation at 95 °C for 45 s, hybridization at the appropriate temperature (Table 1) for 45 s, elongation at 72 °C for 45 s and a final extension stage at 72 °C for 10 min. In addition to the INIA-CGC accessions, positive controls (EA67 and S.288) were included in the screening for all markers.
The PCR products were separated by Capillary electrophoresis (CE). The reaction mix for CE for every sample was 9.7 µl of Hi-Di Formamide, 0.3 µL of GeneScan 600 LIZ Size Standard (Applied Biosystems) and 0.8 µL of each product. The samples were denatured for 3 min at 95 °C, and then cooled to -20 °C for 3 min. The CE was performed on an ABI Prism 3130XL Genetic Analyzer® (Applied Biosystems).
Data analysis
For the representation of the coffee varieties from INIA-CGC was used “ggplot2” R package (Wickham, 2016). The electropherograms were analyzed with GeneMapper v4.0 (Applied Biosystems). Nonspecific products and false positive spikes were removed, as indicated in the user guide. Possible allele size parameters for each molecular marker were established. Clean data was exported as tabulated text and analyzed with “adegenet” package (Jombart, 2008) for the allelic frequency; the function “dist.genpop” for calculate the genetic distance; and the “dendextend” package (Galili, 2015) for dendrogram graphic. All packages mentioned were used in Rstudio software. The scripts used were based on Herrera (2019).
3. Results and discussion
DNA isolation
The CTAB method used for DNA extraction was successful in most of the accessions, the average concentration per sample was 1,138.67 ng/µL and a spectrophotometric 260/280 ratio of 2.10. Lashermes extraction method was used only in those samples with a high concentration of polyphenols and carbohydrates, the average concentration obtained was 298 ng/µL with a 260/280 ratio of 1.6. Two accessions (CA-116 and CA-117) showed low DNA quality with both the CTAB and Lashermes methods, therefore, they were not included in further analysis.
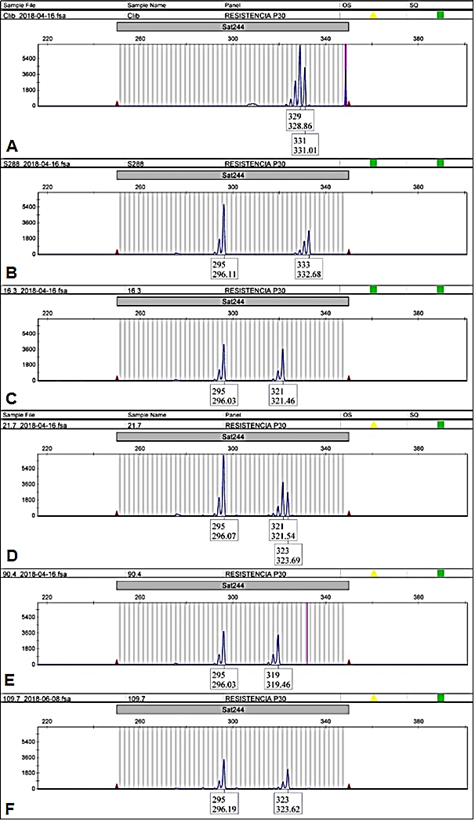
Figure 2 Sat244 electropherogram of EA67, C. liberica (A); S.288, C. arabica x C. liberica (B) and four accessions of INA-CGC: CA-16.3 (C); CA-21.7 (D); CA-90.4 (E) and CA-109.7 (F). The accessions CA-16.3; CA-21.7; CA-90.4 were samples with different genotype from CA-109.7, (295/323) that was the genotype most common in INIA´s Coffee Germplasm Collection.
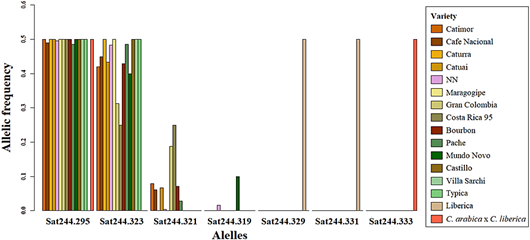
Figure 3 Allelic frequency of Sat244 marker in all samples represented by varieties. Variety denomination according to producers.
The CTAB method was effective for obtaining high-quality DNA from 80% of the processed samples. The remaining samples were rescued using the method developed by Dr. Lashermes. The main difficulty found on sample processing was the presence of mucilage and dark pigments in the DNA pellet that could affect its quality. Inglis et al. (2018) showed that samples with a high number of metabolites and polyphenols impair the quality of DNA extraction. To improve extraction quality, Dr. Lashermes modified the original extraction protocol and used MATAB, sarkosyl and sorbitol to remove polysaccharides (Azmat et al., 2012; Souza et al., 2012), and sodium bisulfite to prevent the action of nucleases during extraction (Byrne et al., 2001). This resulted in higher DNA purity but lower yield. However, for PCR-based genotyping methods, it is always preferable to have higher purity instead of higher yield.
Marker Amplification and capillary electrophoresis
A total of 167 accessions from the INIA-CGC were analyzed. Three biological replicates were amplified for 16 accessions (CA-1 to CA-16); 2 biological replicates for 136 accessions (CA-17 to CA-160) and 1 replicate for 15 accessions (CA-161 to CA-169) making a total of 335 samples processed.
Accessions of INIA-CGC were polymorphic with Sat 244 marker (Appendix). The collection showed 4 different alleles and 5 different genotypes for Sat244 marker (295 bp, 319 bp, 321 bp, and 323 bp; Figure 2). The most common genotype was 295/323 (303 samples, Figure 2F), followed by genotype 295/321 (21 samples, Figure 2C) and genotype 295/319 (5 samples, Figure 2E). Moreover, this marker displayed triallelic genotypes 295/321/323 (in 3 samples: CA-21.7, CA-47.3, CA-107.4, Figure 2D) and genotype 295/319/323 (in CA-97.5, figure not shown). Some samples of the biological replicates produced different genotypes among them (3.5% of the accessions evaluated), e.g., the CA-29 and CA-144 accessions showed genotypes 295/323 and 295/321. Of these alleles, only the 295 bp allele was shared with the hybrid S.288. The positive controls had exclusive alleles 329 bp, 331 bp (EA67, Figure 2A) and 333 bp (S.288, Figure 2B). Furthermore, the allelic frequency of Sat244 showed that the accessions with variety denomination Costa Rica 95 and Gran Colombia had the allele 321 bp, and Mundo Novo the allele 319 bp (Figure 3).
The other three markers (BA-124-12K-f; SP-M8-SH3 and BA-48-21O-f) were monomorphic for the INIA-CGC accessions. Only one allele (403 bp) was observed for BA-124-12K-f; alleles 253 bp and 259 bp for SP-M8-SH3 and the allele 332 bp for the BA-48-21O-f marker.
In all cases, the EA67 control (C. liberica) exhibited unique alleles different from those in the INIA-CGC (allele 349 bp for BA-124-12K-f and allele 257 bp for SP-M8-SH3, showed in Figure 4Aand5A, respectively). The hybrid S.288 control exhibited two alleles in all cases, one shared with the collection and the other shared with EA67 (349 bp and 403 bp for BA-124-12K-f; 253 bp and 257 bp for SP-M8-SH3, showed in Figure 4Band5B). Both positive controls did not amplify with BA-48-21O-f marker.
A variety dendrogram (Figure 6) based on the allelic frequency was obtained by the Nei’s distance (Nei, 1978) and constructed through UPGMA clustering method. Varieties Costa Rica 95 and Gran Colombia (Variety denomination according to producers) were clustered in the same group. Moreover, the dendrogram showed the separation of Liberica (EA67); C. arabica x C. liberica (S.288) and all INIA-CGC accessions.
Molecular markers have been widely used to study genetic diversity of several species (Mohamed et al., 2019; Lee, 2019; Cueva-Agila et al., 2019), including coffee (Mishra, 2020). Moreover, molecular markers have been used to assist selection for agronomic characteristics (Pruvot-Woehl et al., 2020; Sousa et al., 2019; Alkimim et al., 2017; Nsabiyera et al., 2016; Mahé et al., 2008).
INIA-CGC is in the early stages of becoming a formal collection. For this reason, information about the accessions is still in process of manual curation that will include DNA fingerprinting and large-scale phenotyping. In this study, the marker Sat244 was able to discriminate between re plicates (3%) of the accessions screened. Further genoty ping with other microsatellites will assist in the identification of replicates that require reclassification. The use of novel genotyping techniques will facilitate curation by making it more reliable and reproducible. The capillary electropho resis standardized in this study is a good example of accurate and reproducible genotyping and will allow the screening of the collection and larger populations with other microsatellites already described in the literature (Sánchez et al., 2020; Andrianasolo et al., 2013).
The 4 markers used in this study were described by Mahé et al. (2008), these markers have been classified in SSR markers, SCAR markers obtained by bacterial artificial chromosome (BAC) genomic library and a SCAR from AFLP markers identified by Prakash et al. (2004). These markers were evaluated in two populations: a Matari x S.288 F2 mapping population and a backcross (BC2) [(Matari x S.288) x Matari] x Matari (Mahé et al., 2008; Prakash et al., 2004; Prakash et al., 2002). In this work, three of the four markers were useful to differentiate C. liberica control (EA67) from the hybrid C. arabica x C. liberica control (S.288) and the accessions from INIA-CGC.
Tornincasa (2008) used the Sat244 marker, named SAT-01-G10 (that had the same sequence), for variety analyses in Coffea arabica. In her work indicated that SAT-01-G10 could improve the process of variety identification of samples with a non-specific profile of the microsatellite’s markers selected. Therefore, Sat244 marker clustered some C. arabica varieties of INIA-CGC as shown in Figure 6. The polymorphism generated by Sat244 in C. liberica plants was also reported by Mahé (2007); González-Martínez et al. (2009) and Alkimim et al. (2017) that observed different bands profiles in breeding population of C. liberica. Limited information related to the molecular weights of the different alleles of Sat244 in C. liberica, makes it difficult to identify the one that co-segregates with SH3 most frequently.
Surprisingly, the BA-48-21O-f marker could not be amplified in either positive controls, but it was amplified in the INIA-CGC. This marker was developed and evaluated in S.288 and in various mapping populations derived from S.288. Although the linkage map suggests that BA-48-21O-f is the most distant marker from SH3 gene (Lashermes et al., 2010; Mahé et al., 2008), this does not explain the absence of an amplification product in S.288. The amplification conditions were similar for both, INIA-CGC samples and the two positive controls, which rules out the possibility of human errors. To dismiss any possible problems with capillary electrophoresis, the PCR products were also separated on 2.5% agarose gels (data not shown). As a marker belonging to the C. arabica genome (Ca subgenome), BA-48-21O-f could be close to the introgression / recombination point of the SH3 locus. This would explain the absence of product in the EA67 control. For S.288, a possible determining factor was its DNA quality, which exhibited very low 260/280 and 230/260 ratios (data not shown), and agarose gel electrophoresis showed DNA degradation that could have affected the marker biding site. These factors could explain the absence of the BA-48-21O-f marker in S.288.
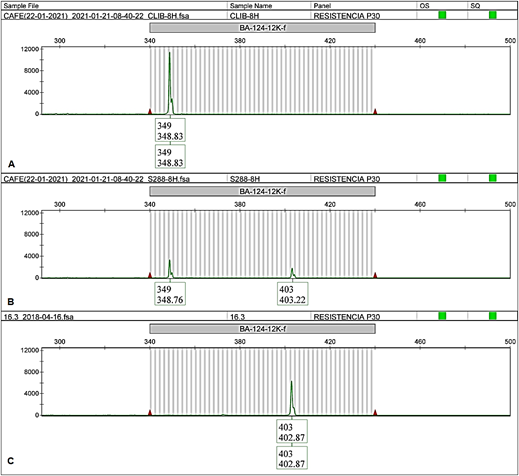
Figure 4 BA-124-12K-f electropherogram of EA67, C. liberica (A); S.288, C. arabica x C. liberica (B) and CA-16.3 from INA-CGC (C).
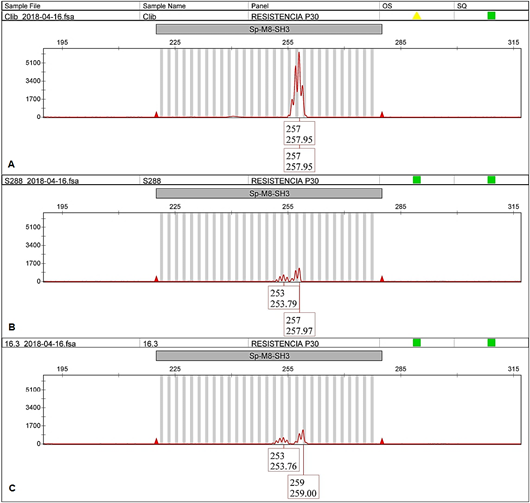
Figure 5 SP-M8-SH3 electropherogram of EA67, C. liberica (A); S.288, C. arabica x C. liberica (B) and CA-16.3 from INA-CGC (C).
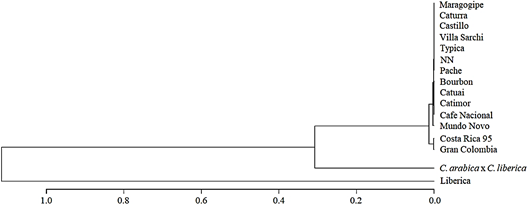
Figure 6 Variety dendrogram by Nei’s distance and constructed through UPGMA clustering method. Variety denomination according to producers.
The BA-124-12K-f marker was used by Alkimim et al. (2017), Valencia et al. (2017) and González-Martínez et al. (2009). Valencia et al. (2017) only used BA-124-12K-f for their SH3 screening in introgressed progenies and concluded that their evaluation can be useful for early selection of plants that carries SH3 gene.
The SP-M8-SH3 marker was selected for screening the INIA-CGC accessions due to its proximity to the SH3 gene, as shown in the linkage map done by Mahé et al. (2008). The results of this study suggest that the best markers to determine the presence of the SH3 gene in coffee collections are BA-124-12K-f and SP-M8-SH3 that clearly discriminate INIA-CGC accessions from the hybrid S.288 and from C. liberica (EA67).
Taken together, these results demonstrate that none of the INIA-CGC accessions possess the SH3 gene that confers resistance to Hv. There are internal reports of tolerance/resistance to Hv in accessions from the INIA-CGC. If this tolerance/resistance is confirmed that it was not introgressed from C. liberica.
4. Conclusions
This is the first study in Peru to demonstrate the usefulness of capillary electrophoresis to evaluate markers linked to the SH3 gene which confers resistance to coffee leaf rust (Hemileia vastatrix) in 335 samples representing 167 accessions of the INIA-CGC. This work will facilitate the use of marker-assisted selection (MAS) in a massive way in large coffee populations and will accelerate the molecular characterization of coffee germplasm to identify samples carrying the SH3 gene that could be used in breeding programs as a source of potential resistance.