1. Introduction
Cassava (Manihot esculenta Crantz) is a resilient crop whose roots have a wide range of uses including human consumption, biofuel and animal feed. A diverse family of viruses can cause disease in cassava worldwide. Among these, Cassava Frogskin Disease (CFSD) is the most economically important disease of cassava in the Latin America region (Pardo et al., 2022). There are official reports of CFSD in Brazil, Colombia, Costa Rica, Paraguay, and Venezuela. (Pineda et al., 1983; Álvarez et al., 2009; Cardozo et al., 2016; Chaparro-Martinez & Trujillo-Pinto, 2001; de Oliveira et al., 2020). Yield losses due to the disease range between 10 and 90 %, because it directly affects starch accumulation (Pardo et al., 2022). The main causal agent of CFSD is yet to be identified as there are several pathogens detected in affected plants (Alvarez et al., 2009; Calvert et al., 2008; Carvajal-Yepes et al., 2014; de Oliveiraet al., 2020).
Cassava yields can be severely reduced by accumulation of pathogen infections during vegetative crop propagation, a phenomenon also known as seed degeneration (Thomas-Sharma et al., 2017). Viruses are among the most severe cassava pathogens worldwide (Legg et al., 2015; Lozano et al., 2017; Siriwan et al., 2020; Collavino et al., 2021). In Africa, viruses belonging to the Geminiviridae and Potyviridae families predominate, and cause severe diseases, while in the Americas, where no cassava-infecting geminivirids have been reported, viruses of the Alphaflexiviridae family (Calvert et al., 1996; Lozano et al., 2017) are the cause of significant yield reductions in cassava (Venturini et al., 2016; Collavino et al., 2021). Novel cassava-infecting viruses in the Americas have been recently reported in plants affected by CFSD (Carvajal-Yepes et al., 2014). This is a regionally endemic disease of cassava, characterized by longitudinal lip-like fissures, and the root peel presenting a thick, cork-like appearance (Pardo et al., 2022). Among these viruses, a secovirid was shown to cause clear leaf symptoms and was found most commonly in mixed infections, in all regions where CFSD is reported (Carvajal-Yepes et al., 2014; Jimenez, 2017; de Oliveira et al., 2020).
The family Secoviridae is the only one in the order Picornavirales containing plant-infecting viruses and within it, the genus Torradovirus is a group of newly-emerging plant viruses, identified mostly via high-throughput sequencing (HTS) technologies (van der Vlugt et al., 2015). Torradoviruses were initially identified via classical technologies in tomatoes, where they cause characteristic symptoms of necrotic spots on leaves surrounded by yellow areas, growth reduction and severe necrotic rings on fruits (Verbeek et al., 2007). Several other torradoviruses have been identified via HTS in other hosts (van der Vlugt et al., 2015). The torradovirus we report here was temporarily named Cassava ‘torrado-like’ virus (CsTLV), but it contains all the hallmarks found in members of the torradovirus genus (Van der Vlugt et al., 2015; Jimenez, 2017; Leiva et al., 2021). Torradoviruses are small spherical virions of around 30 nm in diameter composed of three coat proteins: Vp35, Vp26 and Vp23 which encapsidate a bipartite single-stranded positive-sense RNA genome [ss(+)RNA] (Verbeek et al., 2007). RNA1 size ranges from 6 911 to 7 802 nucleotides (nt) long and RNA2 from 4 701 to 5 695 nt (van der Vlugt et al., 2015). A distinct characteristic of torradoviruses is the presence of an overlapping ORF at the 5’ end of RNA2, which encodes for a, ~25 kDa, which in tomato apex necrosis virus (ToANV) is involved in long-distance movement and essential for systemic infection in Nicotiana benthamiana (Ferriol et al., 2018). This characteristic prompted us to hypothesize a role for this protein in virus movement. However, in the absence of a virus infectious clone for cassava, our approach was to use Agro-infiltration to express the test gene in plants, which are later mechanically infected by a virus that is not as efficiently transmitted.
Virus movement into plants to establish a systemic infection is divided into two main steps: i) local movement from cell to cell through plasmodesmata, and ii) long-distance movement via the phloem (Kumar & Dasgupta, 2021). During mechanical inoculations, the virus needs to first enter a cell and replicate. At this stage, the virus must also transit the plasmodesmata, for which the Triple Gene Block (TGB) deployed by the Alphaflexiviridae is one of the most-studied mechanisms (Solovyev et al., 2012). TGB proteins are involved in cell to cell and vascular transport, delivering the genomes via plasmodesmata by increasing the size-exclusion limit, allowing viral spread to surrounding cells. Briefly, the three proteins interact as follows: multifunctional protein TGB1 increases the permeability of plasmodesmata for virus cell to cell movement; and small membrane-associated proteins TGB2 and TGB3 mediate TGB1 transport to the plasmodesmata (Morozov & Solovyev, 2020). Interestingly, cassava virus X (CsVX) is an alphaflexivirid that lacks the TGB3 protein and has a low efficiency (~20%) of mechanical inoculation in N. benthamiana plants (Lozano et al., 2017). Lacking a TGB protein could explain the low efficiency in transmission observed for CsVX, (Verchot-Lubicz, J., 2005) when compared with another alphaflexivirid, cassava common mosaic virus (CsCMV), which contains a complete TGB and exhibits a higher mechanical transmission efficiency (Lozano et al., 2017). Therefore, CsVX was chosen as a model to assay a role for RNA2-ORF1 in virus movement.
2. Materials and methods
2.1 Plant material and virus isolates
N. benthamiana seeds were germinated on moist sterilized soil sand mix (2:1). After 15 days, two young seedlings were transplanted into plastic 10 x 8 cm pots. CsTLV (isolate Sec13, GenBank accession number RNA1, MF449522; RNA2, MF449523) isolated from an in vitro cassava plant genotype Secundina (MCOL2063) (Jimenez, 2017), was later maintained by graft transmission, in the genotype Reina (CIAT accession number CM6740-7), under greenhouse conditions at the International Center for Tropical Agriculture (CIAT) in Palmira, Colombia. This isolate was the source of the CsTLV genes used in this work. CsVX isolate Ven164 (GenBank accession number NC_034375.1), was used as an indicator of mechanical inoculation and has been previously described by Lozano et al. (2017).
2.2 Agroinfiltration constructs for transient expression in N. benthamiana leaves
To functionally characterize CsTLV RNA2-ORF1, we used Agrobacterium-mediated transient gene expression assays. For transient expression we used Agrobacterium tumefaciens strain EHA105. To amplify the coding region of the Maf/HAM1 and RNA2-ORF1 proteins, total RNA extraction from cassava leaves was done using CTAB, and cDNA was synthesized as previously described by Jimenez et al. (2021). To obtain the ORF1-RNA2 and the Maf/Ham1 (used as control) coding sequences, we used primers: CsTLV_RNA2_ORF1_F (5’-AATATGGCGGCCGCGTCTTTCATTGATTCCTTGGA-3’) and CsTLV_RNA2_ORF1_R (5’-AATGGCCGGCCTCATTCTGCTTCAGGGGTTTGAG-3’). CsTLV_RNA1_HAM_F (5’-ATTATGGCGGCCGCGAAGTTTCTGATCACCACCAC-3’) and CsTLV_RNA1_HAM_R (5’-AATGGCCGGCCTCATGTTATCTTACGCAAACCA-3’), respectively. The above primers contained the restriction sites NotI at the 5’ end and FseI at the 3’ end (underlined).
RT-PCR reactions were performed under the following conditions: cDNA was denatured at 94 °C for 5 min, followed by 35 cycles of amplification (94 °C for 45 s, 64.2 °C for 1 min and 72 °C for 1 min) and 10 min at 72 °C. DNA fragments from PCR reactions were gel-purified using an E.Z.N.A. Gel Extraction Kit (Omega) and the plasmids were purified using a Wizard® Plus SV Minipreps DNA Purification System kit (Promega). The constructs were cloned in two steps using three different vectors. The first step consisted of cloning the fragments in pGEM-T easy vector (Promega) and transferring to the pK vector. In the second step, the constructs were transferred from the pK vector to the pA vector, where construct expression is under the control of the cauliflower mosaic virus (CaMV) 35S promoter with spectinomycin-resistance. The restriction fragments obtained from the digestion of the pK-Ham1 and pK-ORF1 plasmid with AscI y SbfI (New England BioLabs Inc.) were subcloned into the pA vector. The resulting plasmids were named pA-Ham1 and pA-ORF1. pA vectors were mobilized from Escherichia coli DH5α into A. tumefaciens EHA105 via electroporation. A. tumefaciens colonies, capable of growing in a spectinomycin-containing medium, were selected for transient expression in N. benthamiana leaves.
2.3 Agro-infiltration and mechanical inoculation in N. benthamiana
The construct pA-GFP (green fluorescent protein) (Kreuze et al., 2005) was used to visualize the transient gene expression in agro-infiltrated cells of N. benthamiana and determine the highest expression time of the transgene. Four N. benthamiana plants were used for agro-infiltration assays per construct. Agro-infiltration assays were carried out at six weeks post-germination, when the plant presented 4‒6 true leaves. One colony of A. tumefaciens containing the constructs was grown in a 100 mL flask for 16 h, at 28 °C using a shaker in 20 mL of LB medium, supplemented with rifampicin [30 μg/mL], spectinomycin [100 μg/mL], MES 10 mM and acetosyringone 20 μM. The next day, cells were harvested by centrifugation at 3 000 rpm for 20 min at room temperature Then, these were resuspended in 2 mL of induction medium (MgCl2 10 mM, MES 10 mM, acetosyringone 150 μM) to a final optical density of 0.5 at 600 nm.
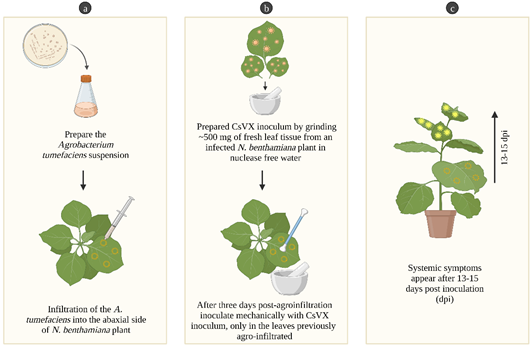
Figure 1 Agrobacterium tumefaciens-mediated transformation and mechanical inoculation in N. benthamiana. a. Preparation of A. tumefaciens suspension and infiltration into the leaves. b. Preparation of CsVX inoculum and mechanical inoculation. c. Systemic symptoms on new leaves. Created with BioRender.com.
Finally, bacterial suspensions were incubated at room temperature for three hours before agro-infiltration. Four zones on the backside of each leaf were agroinfiltrated us ing a needleless syringe and applying gentle pres sure. The effect was monitored for five days, under a 100 W UV lamp (long-wave model B-100AP ‒UVP, Germany).
To study the effect of the expressed construct on the transmission efficiency of CsVX, six N. benthamiana plants were agroinfiltrated as men tioned above, and after three days post-agro-infiltration (dpa) these plants were mechanically in oculated with a CsVX inoculum. The virus inoculum was prepared by grinding ~500 mg of fresh leaf tissue from an infected N. benthamiana plant, in nuclease free water (w/v: 1:3). The first 4-6 true leaves of N. benthamiana were used for mechanical inoculation using cotton swabs and carborundum (Fisher Scientific), as described by Lozano et al. (2017) (Figure 1). The plants were maintained until 30 dpa. As controls, we used plants that were only mechanically inoculated with sterile water and plants that were only mechanically inoculated with the virus. The assay was replicated twice.
2.4 Virus detection
Plants were monitored daily for CsVX symptoms during the 30-day experimental period. Leaf symptoms were recorded by photograph. We collected the 3‒4 top youngest leaves at 20 and 30 dpa, wrapped them in Kimwipes® and stored them in Ziploc® bags containing silica gel (Merck), until processing as previously reported (Jimenez et al., 2021). CsVX was detected using a 1:20 proportion (~14 mg) of dried leaf tissue by double antibody sandwich-enzyme-linked immunosorbent assay (DAS-ELISA), as described by Lozano et al. (2017).
3. Results and discussion
3.1 Considerations of CsTLV as an atypical member of the Secoviridae family
CsTLV is a novel bipartite member of the Torradovirus genus (Figure 2b), reported in mixed infections in cassava plants displaying CFSD root symptoms in Colombia and Brazil (Carvajal-Yepes et al., 2014; de Oliveira et al., 2020). CsTLV induces leaf chlorotic spots symptoms in Secundina and Reina, around four weeks after graft inoculation under greenhouse conditions (Figure 2a).
CsTLV is an atypical member of the Secoviridae, containing a Maf/Ham1 domain located in RNA1 downstream of the RdRp domain (Leiva et al., 2022). Maf/HAM1 proteins are nucleoside triphosphate (NTP) pyrophosphatases that protect cells by hydrolyzing non-canonical NTPs, preventing their incorporation into DNA or RNA (Noskov et al., 1996; Mbanzibwa et al., 2009; Tomlinson et al., 2019; Valli et al., 2022). Recent results demonstrated that Maf/Ham1 domain is associated with the development of necrosis in N. benthamiana plants (Tomlinson et al., 2019) and Valli et al. (2022) suggests that the HAM1 domain participates in viral replication because it might be covalently bound during CBSV and EuRV infections. Potential roles in infection and possible functions of the Maf/Ham1 domain are discussed by James et al. (2021). Preliminary results indicate that it has no role in RNA silencing (Mbanzibwa et al., 2009; Jimenez, 2017), and in the assays presented here, we did not find a role in increasing the efficiency of transmission by mechanical inoculation (virus local movement) of CsVX in N. benthamiana.
RNA2-ORF1 of the CsTLV is 678 nt long and encodes a predicted protein of 226 aa (~25 kDa) (Figure 2b). This gene is conserved in all torradoviruses and is closely related to its homologue in squash chlorotic leaf spot virus (SCLSV, GenBank acc number YP_009389537) (Figure 3a). BLASTp analysis of this gene revealed a highest aa sequence-identity of 31% (coverage of 99%) between CsTLV and SCLSV. Sequence alignment showed two conserved motifs of RNA2-ORF1. Interestingly the conserved torradovirus LDF motif is absent in CsTLV and is closest relative SCLSV. In contrast the highly conserved PQ motif, is present in all torradoviruses and located at position 68‒69 in CsTLV-Sec13 (Figure 3b) (van der Vlugt et al., 2015).
3.2 Efficiency of mechanical inoculation and symptoms detection in agroinfiltrated plants
Secoviridae is the only family in the order of Picornavirales that contains plant-infecting viruses. Two secovirids are reported infecting cassava: cassava green mottle virus (CsGMV; Lennon et al., 1987), from the Solomon Islands and cassava American latent virus (CsALV; Walter et al., 1989), reported in South America. Although the complete genome sequences of these viruses are not yet available to establish their genetic differences, a clear biological difference with CsTLV is that both, CsGMV or CsALV can be mechanically transmitted to N. benthamiana and other hosts, while CsTLV could not be transmitted to a range of alternative hosts including N. benthamiana, Chenopodium amaranticolor, Physalis sp., Datura stramonium, Solanum lycopersicum var. Manalucie, Gomphrena globosa, Nicandra physaloides, Physalis ungulata, Nicotiana clevelandii and Nicotiana tabacum var. Samsun NN (Jimenez, 2017).

Figure 2 Foliar symptoms induced by CsTLV isolate Sec13 in cassava plants and genome organization. a. Leaf chlorotic spots symptoms observed in the genotypes Secundina and Reina. b. Schematic representation of a generic torradovirus genome showing the conserved sequence motifs and the location of RNA2-ORF1 (yellow box). Legend: Hel: helicase, PRO: 3C-like protease, RdRp: RNA-dependent RNA polymerase, A3: movement protein, CP: coat protein.
Based on these previous observations, the delivery of CsTLV proteins to evaluate their functions was carried out through agro-infiltration, using GFP as a marker of gene expression.
GFP fluorescence in agro-infiltrated N. benthamiana leaves observed under UV light was detected at 3 dpa and faded out by 5 dpa, therefore, we carried out the mechanical inoculation of CsVX in leaves that were agro-infiltrated at 3 dpa with the constructs expressing the CsTLV proteins Maf/Ham1 and RNA2-ORF1. As previously reported by Lozano et al. (2017), CsVX isolate Ven164 did not show visual symptoms in cassava in single infections (not shown), but the virus caused systemic infections and typical symptoms in the indicator plant N. benthamiana, as observed here (Figure 4). The symptoms were yellow spots with irregular borders that subsequently became necrotic at 20‒25 days post-inoculation (dpi). These N. benthamiana plants were used as inoculum and in general, we obtained a 33.3 % efficiency of mechanical transmission to N. benthamiana (Figure 4b).
In contrast, the alphaflexivirid CsCMV has a complete TGB and a 90% ‒ 100% efficiency of mechanical transmission (Lozano et al., 2017). Reported roles for TGB3 in virus infection are in cell-to-cell movement and as a symptom determinant, as evaluated in Chenopodium quinoa plants infected by bamboo mosaic virus (Wu et al., 2011) and N. benthamiana infected by potato virus X (Tamai & Meshi, 2001).
All plants that presented characteristic symptoms of CsVX in N. benthamiana were positive to the ELISA test, meanwhile, one plant without clear symptoms of CsVX also was positive. Curiously, this asympto matic plant belonged to the Maf/Ham1 treatment and until 30 dpa it remained asymptomatic, and although it was considered as positive for systemic infection, it reached only 33.3% of systemic infection for the Maf/Ham1 treatment. None of negative control plants gave positive reactions in ELISA tests, indicating that there was no cross-contamination among treatments. Additionally, a two-factor with-replication ANOVA test found no significant differences (F= 0.00827, F crit = 3.986269, P-value= 0.927) in the average titers of CsVX among the inoculated plants, among treatments between 20-30 dpa. This indicates that the virus reaches its highest concentration in symptomatic leaves at 20 dpa or before (Figure 4a). Furthermore, there were differences in the time in which the plants started to showed symptoms. Plants previously agro-infiltrated with RNA2-ORF1 show symptoms 2‒4 days earlier than other treatments. On average, systemic virus symptoms in N. benthamiana appeared at 11 dpi in plants that were agro-infiltrated with the pA-ORF1 construct.

Figure 3 Phylogenetic relationships of RNA2-ORF1 CsTLV-Sec13. a. Phylogenetic relationships of CsTLV with members of the torradovirus genus using the aa sequence of RNA2-ORF1. The phylogenetic tree was generated by Neighbor-Joining using MEGA v6. The evolutionary distances were computed using the Poisson correction method and are in the units of the number of a.a. substitutions per site. b. Sequence alignment showing the conserved amino acids shared (black asterisks), the black bar indicates the location of the conserved LDF motif in all torradoviruses (absent in CsTLV and SCLSV), PQ motif (red asterisks). The graph was plotted using MView (Brown et al., 1998).
Abbreviations: ToMarV, tomato marchitez virus; ToNDV, tomato necrotic dwarf virus; ToChSV, tomato chocolate spot virus; ToTV, tomato torrado virus; ToChV, tomato chocolate virus; CaTV1, carrot torradovirus 1; MYMoV, motherwort yellow mottle virus; RCTV-1, red clover torradovirus 1; LNLCV, lettuce necrotic leaf curl virus; SCLSV, squash chlorotic leaf spot virus; CsTLV, cassava torrado-like virus.
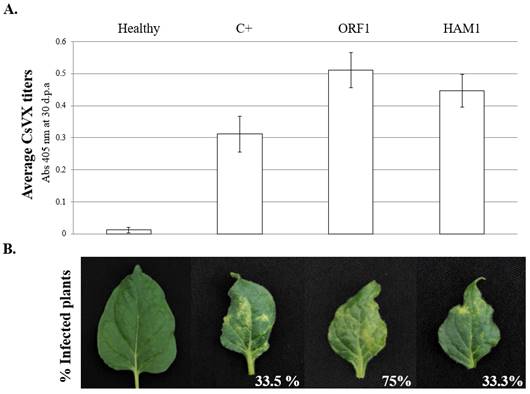
Figure 4 ELISA results and symptoms obtained after agro-infiltration and mechanical inoculation of CsVX in N. benthamiana. a. DAS ELISA titers at 30 dpa. Absorbance values were measured at 405 nm. ANOVA test found no significant differences (P > 0,05) in virus titers among treatments. b. Leaf symptoms of CsVX and effect of RNA2-ORF1 on the infection efficiency in N. benthamiana plants, showing 2.3 fold increase in infection efficiency in plants previously agroinfiltrated with RNA2-ORF1. The error bars represent the standard deviation obtained. Legend: Positive control (C+).
In other treatments, symptoms started to appear at 13‒15 dpi in systemic leaves. Here, plants that did not show symptoms at 10‒20 dpi remained symptomless until the end of the experiment (30 dpa). We did not observe differences in the CsVX titers among symptomatic plants (Figure 4a). Nevertheless, a significant difference was obtained in the percentage of plants infected with RNA2-ORF1 treatment as compared to the other treatments. The average number of plants infected with CsVX and showing disease symptoms was 33.3% in plants previously agro-infiltrated with the Maf/HAM1 expressing construct, and 33.5% in virus-inoculated plants without previous agro-infiltration. In contrast, the average frequency of systemic infection in N. benthamiana plants agro-infiltrated with RNA2-ORF1 was 75% (Figure 4b).
Previous research on RNA2-ORF1 from a tomato-infecting torradovirus ToANV, indicated that this protein is required for systemic virus movement, before uploading of the virus into the phloem, in N. benthamiana and tomato plants (Ferriol et al., 2018). Our results show that the percentage of plants systemically infected with CsVX, was significantly increased (2.3-fold) when plants were previously expressing the RNA2-ORF1. This suggests that, as with the RNA2-ORF1 of ToANV, the homologous gene in CsTLV may have a role in virus movement. Further investigations may link these observations with the rare occurrence of CsVX in single infection and its rather common occurrence in mixed virus infections, in field collected plants (Harrison et al., 1986; Nolt et al., 1992; Lozano et al., 2017).
4. Conclusions
In conclusion, the novel secovirid CsTLV associated with leaf spot symptoms in cassava encodes a gene that could enhance other viral infections in N. benthamiana. Further studies are required to elucidate this effect and its role in mixed infections in cassava often observed in plants affected by CFSD.