1. Introduction
Cocoa (Theobroma cacao L.) is one of the crops of great economic importance in the world. The global chocolate market size was estimated at USD 113.2 billion in 2021. The crop has great agroeconomic importance worldwide due to the high demand for chocolate in the cosmetic and food industries (Motamayor et al., 2013; Garcia et al., 2021). In the future, a shortage of cocoa is expected due to the low production yield of small farmers who are the ones currently fulfilling most of the cocoa market demand, this as a result of fungal and viral diseases, climatic, and soil conditions which severely affect the crop (Arévalo-Gardini et al., 2017; McElroy et al., 2018; Wickramasuriya & Dunwell, 2018).
The biotic and abiotic stressors that affect cocoa have been investigated and understood at the genetic level, through the assembly and different analyses of its genome. Cocoa has a genome of approximately 445 Mbp, with some variations depending on the variety or clone (Wickramasuriya & Dunwell, 2018). Genomics have made it possible to study and elucidate the function of genes of great agronomic value, and through breeding it is expected to generate cultivars of higher quality (da Silva et al., 2017; Abdullah et al., 2021).
Knowledge of the functionality of genes is essential when it comes to performing Cocoa breeding, which can be achieved through gene overexpression, inactivation, silencing and genetic editing, allowing the integration of agronomically important traits. A good example of success of cocoa breeding is the obtainment of varieties with resistance to Colletotrichum gloeosporioides (Maximova et al., 2006; Khalil et al., 2022). The success of genetic modification depends on the regeneration capacity of transformed plants. Somatic embryogenesis of cocoa is still reported as inefficient due to the low rate of embryos produced and lack of maturity. These traits are genotype-dependent, whereas the cocoa crop is recalcitrant under in vitro conditions (Maximova et al., 2002; Traore & Guiltinan, 2006; da Silva et al., 2008).
An alternative solution can be found in the study of transcription factor (TF) families, which are proteins that regulate gene transcription, thus presenting activator TFs, which promote the transcription of a gene, while repressor TFs decrease transcription (Carrillo et al., 2023, Kajla et al., 2023; Samtani et al., 2023; Zou & Sun, 2023). Within the Arabidopsis thaliana gene family, the TF BABY BOOM (BBM) was differentiated with a high degree of similarity to the corresponding gene in cocoa (Florez et al., 2015, Pierroz, 2023). This possibility promotes the formation of somatic embryos through plant cell regulation of the totipotency by ectopic overexpression being introduced to the plant by transformation as a TF for posthumous induction (Horstman et al., 2017, Ahn et al., 2023).
Agroinfiltration is considered one of the most efficient transformation techniques, which involves the use of the bacterium Agrobacterium tumefaciens. This bacterium infects the plant by transferring the tumor-inducing (Ti) DNA into the genome of the plant through the chemical stimuli released by wounds on the leaves. The Ti-DNA is modified to create a cloning vector into which the gene of interest can be inserted for integration into the plant genome. Tzfira et al. (2002) have extensively researched and documented this process. When a TF is introduced, it is typically used to study gene expression and functionality in plants. This makes it a useful tool for identifying valuable genes that can be improved genetically through transient expression, as highlighted by Guidarelli & Baraldi (2015).
Although attempts have been made to use this technique for inducing embryogenic calluses in Peruvian cocoa leaf tissue, few studies have been conducted due to the tissue’s recalcitrant nature. However, its accessibility for laboratory manipulation makes it a promising candidate for research purposes, as suggested by Shires et al. (2017). Therefore, the current study aims to genetically transform the transcriptional factor BABY BOOM of Arabidopsis thaliana (AtBBM) to induce the formation of embryogenic calluses in leaves of elite cocoa plants IMC-67.
2. Methodology
Plant material
Arabidopsis thaliana L. seeds ecotype Columbia (Col-0) was established under aseptic conditions (in vitro) in the laboratory of Biología y Genética Molecular de la Universidad Nacional de San Martín (UNSM), Tarapoto, Peru. The seeds of Arabidopsis thaliana were grown in MS culture medium (Murashige & Skoog, 1962) at half their strength, supplemented with 10 g/l of sucrose, 4 g/l of agar and with pH 5.75. The plant material was kept under controlled conditions (19 °C temperature, 18 hours of light and 6 hours of darkness), until the seedlings presented root systems of approximately 5 cm.
RNA isolation and AtBBM gene amplification
Seedlings of A. thaliana obtained under aseptic conditions were used for RNA extraction using the reagent Trizol and PureLink RNA Mini Kit (Omega Bio-tek, Norcross, GA, USA), following the manufacturer's instructions. The roots of the seedlings were excised and thawed in liquid nitrogen. The RNA samples obtained were quantified, and their quality was assessed using a NanoDrop 2000c (Thermo Scientific). Finally, the quality of the extracted RNA was evaluated by electrophoretic run in 1% agarose gel stained with SYBRsafe.
Reverse transcription was performed using the GoScript Reverse Transcriptase kit (Promega, Madison, WI, USA) to amplify the AtBBM gene fragment with the primers AtBBM-Fv (5′-CACCATGAACTCGATGAATA-3′) and AtBBM-Rv (5′-CTAAGTCGTTCCAAACTGAA-3′) with the enzyme Phusion DNA Polymerase under the following conditions: 98 °C for 30 s; 35 cycles at 98 °C for 30 s, 56 °C for 30 s, 72 °C for 2 min, followed by a final extension period at 72 °C for 10 min. The PCR products were visualized on 1% agarose gel. The amplified fragment was then purified with the Illustra GFX PCR DNA and Gel Band Purification Kit (Cytiva).
Vector construction and cloning
Ligation of the AtBBM fragment (amplified and purified in the previous process) and with the intermediate vector pENTR/D-TOPO (Invitrogen) was performed as established in the protocol of the pENTR™/D-TOPO™ Cloning Kit (Invitrogen). The resulting construct was then transformed into chemically competent Escherichia coli DH5α cells. Once transformed, the bacteria were cultured on plates containing Luria Bertani (LB) medium with kanamycin (50 mg/l) and incubated at 37 °C for 16 h. Subsequently, the grown colonies were picked, and the insert was amplified via PCR with the primers M13-Fv (5′-GTAAAACGACGACGGCCAG-3′) and AtBBM-Rv (5′-CTAAGTGTCGTTCCAAACTGAA-3′), under PCR conditions described above. PCR prod ucts were visualized on 1% agarose gel. Plasmid DNA was extracted from positive colonies using the GenElute Plasmid Miniprep Kit (Sigma Aldrich). The extracted plasmid DNA was sent for sequencing to Macrogen Korea (Seoul, South Korea) using the pri mers M13Fv and M13Rv (5′-GCGGATAACAATTTCACACACACAGG-3′). The obtained sequences were analyzed and compared with the AtBBM sequence described by Florez et al. (2015).
The recombination of the pENTR/D-TOPO vector with the AtBBM gene and the pK7WG2 target vector with the LR Clonase II Kit was carried out according to the manufacturer's protocol. This process involved transforming the construct into chemically competent E. coli DH5α cells, which were grown overnight in LB medium with 50 mg/l of kanamycin. The resulting colonies were then analyzed by PCR to confirm the presence of the pK7WG2::AtBBM construct, using the primers 35s-Fv and AtBBM-Rv under the PCR conditions and plasmid DNA was ex tracted from positive colonies as described above.
The pK7WG2::AtBBM constructs were introduced into Agrobacterium tumefaciens strain GV3101 by electroporation using an electrical pulse of 2.2 KV. The resulting inoculum was then extended on solid LB medium with 50 mg/l of spectinomycin and 50 mg/l of gentamicin and incubated at 28 °C for 48 hours. To confirm the presence of the vector, DNA was extracted from surviving colonies, and PCR was performed using the primers M13-Fv and AtBBM-Rv under the PCR conditions described above.
Transformation of cocoa leaves mediated by Agrobacterium
The transformation of cocoa leaves was developed using the protocol established by Fister et al. (2018), with modifications. In this study, the method of introducing Agrobacterium tumefaciens was tested using the vector pK7WG2::AtBBM, and the coculture of Agrobacterium tumefaciens was tested with and without vacuum infiltration, along with an absolute control (no vacuum/no bacteria).
One day prior to transformation, cultures of Agrobacterium tumefaciens GV3101 were grown with the vector pK7WG2::AtBBM in medium 523 (Viss et al., 1991) with 50 mg/L of kanamycin, and were incubated at 28 °C in darkness with 200 rpm for 16 hours. After 16 hours, the cell density was evaluated and adjusted to an optical density (OD600) of 1.0 using 523 medium. Samples were centrifuged at 1500 rpm for 15 minutes to settle the bacteria and were resuspended with induction medium (Fister et al., 2018). The resuspended bacteria were stirred at 100 rpm at 25 °C for 5 hours in darkness. After the estimated time, pure Silwet L-77 was added to the Agrobacterium culture at a final con centration of 0.02%. The solution was placed in a petri dish and transferred to a desiccator connected to a vacuum pump. Sections of type C cocoa leaves obtained from elite IMC-67 cocoa plants belonging to the nursery of the Laboratorio de Biología y Genética Molecular were placed in the solution.
Transformation by vacuum infiltration was per formed at a pressure of −22 in. Hg for 20 minutes for each side of the leaf sections (abaxial and daxial), and treatments without vacuum filtration (including control) were maintained for the same period in the solution. Subsequently, the leaf sections were removed and placed on sterile filter paper to remove excess bacteria. The leaf sections were then placed with coculture in ED4 medium (García et al., 2018) without antibiotics for 48 hours at 25 °C in total darkness. After 48 hours, the cocoa leaf sections were transferred to ED4 medium with 200 mg/L moxalactam to eliminate Agrobacterium and 50 mg/L of kanamycin as a selection agent, with the conditions of 25 ° C in darkness. The medium was refreshed every 14 days with 50 mg/L of kanamycin, on four occasions, and was maintained under the same conditions as described above.
Molecular analysis of cocoa transforming leaves
Genomic DNA was isolated from cocoa leaf tissue using automated DNA extraction equipment KingFisher™ Duo Prime Purification System (Thermo Fisher Scientific) using the MagMax Plant DNA isolation kit (Applied Biosystems), following the manufacturer’s instructions. To confirm the presence of the AtBBM transgene previously de scribed, PCR assays were performed with the primers AtBBM-Fv and AtBBM-Rv under the PCR conditions described above. The products were displayed in 1% agarose gel.
Statistical Analysis
An experimental design based on a completely randomized design (CRD) was employed to evaluate embryogenic callus formation in cocoa leaves and transformation efficiency in cocoa leaf explants. Each experimental unit consisted of a plate containing 10 leaf fragments obtained from elite IMC-67 cocoa plants, and five replicates were performed for each treatment. The data obtained were analyzed using analysis of variance (ANOVA) and the means were compared using Tukey's test with a significance level of p < 0.05. The statistical analyses were performed using R software (version 4.1.3) (R Core Team, 2020).
3. Results and discussion
Isolation and characterization of the AtBBM sequence
The amplification of the AtBBM candidate gene yielded DNA amplicons ranging from approximately 1500 to 2000 bp in size (Figure 1A), consistent with the expected amplicon size for the primers used. To confirm that the PCR product was indeed the target gene fragment, the DNA amplicons were sequenced, and a 1755 bp DNA sequence was obtained (Figure 1B). The DNA sequence showed a high degree of homology (100%) with the Arabidopsis thaliana group, the integrase-like DNA-binding superfamily protein (EF687843.1), also known as the AtBBM gene.
Sequencing of the coding region of the transcription factor BABY BOOM (AtBBM) isolated from Arabidopsis thaliana root tissue has the potential to compare orthologs in other species. The high homology of AtBBM with the transcription factor BABY BOOM in Theobroma cacao (TcBBM) suggests that they are related and can regulate the growth and development of somatic embryos by transcribing target genes (Florez et al., 2015; Keilwagen et al., 2016). AtBBM can be utilised for genetic transfor mation research in different crops, as reported in studies that examine the effects of heterologous expression of Arabidopsis BBM genes (AtBBM) on callus formation in tobacco (Nicotiana tabacum L.) (Srinivasan et al., 2006).
Vector construction and cloning
The formation of individual bacterial colonies is an indication of successful bacterial transformation using recombinant plasmids. In the case of the plasmid pENTR/D-TOPO containing the AtBBM gene, several individual bacterial colonies were observed (Figure 2A). The validation was then carried out using specific primers for the AtBBM gene, and the positive bacterial colonies showed DNA bands of 1789 bp (Figure 2B). These results confirm that the plasmid successfully transformed into competent bacterial cells is a recombinant plasmid, which means that the AtBBM gene has been successfully integrated into the pENTR/D-TOPO cloning plasmid.
The fragment of the AtBBM gene was subcloned into the expression vector pk7WG2 using ligase enzymes, resulting in the pk7WG2::AtBBM construction cassette. Successful subcloning was confirmed by PCR validation with M13 primers, which produced 1875 bp DNA bands (Figure 3A). The pk7WG2::AtBBM construction cassette includes the AtBBM gene as well as the kanamycin antibiotic gene marker for plant selection (Figure 3B). The AtBBM gene in this construction cassette is regulated by the constitutive cauliflower mosaic virus (CaMV) 35S promoter and terminator.
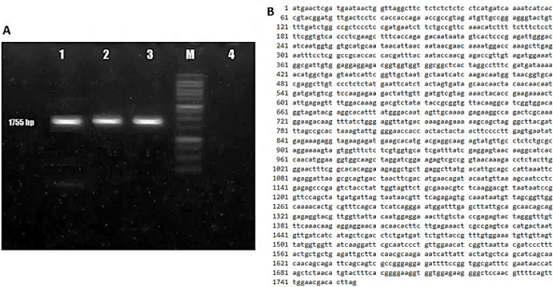
Figure 1 Isolation and characterization of the BBM gene sequence of Arabidopsis thaliana. (A) PCR amplification product of 1755 bp using AtBBM-specific primers with cDNA template (lane 1-3), DNA molecular weight marker - 100 bp DNA Ladder (M), negative control (lane 4). (B) Results of nucleotide sequence analysis of amplicons of the candidate AtBBM gene.
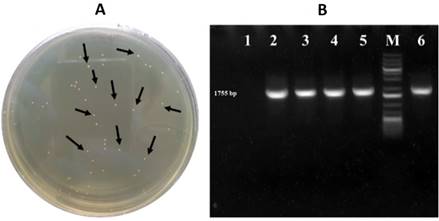
Figure 2 Cloning AtBBM in the pENTR/D-TOPO vector. (A) Positive colonies of Escherichia coli DH5 in solid LB medium transformed with the recombinant plasmid pENTR/D-TOPO::AtBBM (black arrows). (B) PCR amplification product of 1755 bp individual colonies of E. coli DH5 transformed with the plasmid pENTR/D-TOPO::AtBBM (lane 2-5), DNA molecular weight marker - 1 Kbp DNA Ladder (M), Negative control (lane 1), Positive control (lane 6).
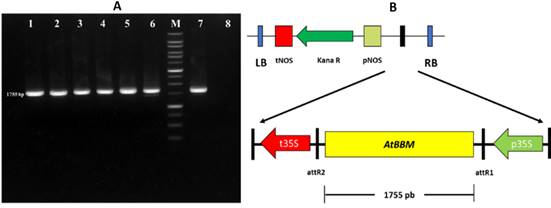
Figure 3 AtBBM cloning in the pk7WG2 expression vector. (A) PCR amplification product of 1755 bp individual colonies of Agrobacterium tumefaciens GV3101 transformed with the plasmid pk7WG2::AtBBM (lanes 1-6), DNA molecular weight marker - 1 Kbp DNA Ladder (M), positive control (lane 7), negative control (lane 8). (B) Map of the plasmid of overexpression pk7WG2::AtBBM.
The use of the Gateway system in vector construction provides several advantages, such as the ability to maintain and reproduce the isolated gene when introduced into the pENTR/D-TOPO cloning plasmid. Additionally, the kanamycin resistance selection gene in the pk7WG2::AttBBM expression plasmid allows easy discrimination of putative calluses that may indicate successful transformation, as plants are sensitive to kanamycin in tissue culture. The presence of the constructed cassette was later confirmed by PCR (Akter et al., 2012; Santoso et al., 2018; Ontiveros-Cisneros et al., 2022). However, it has been reported that the use of kanamycin in other transgenic crops can result in a higher proliferation of calluses that do not regenerate. Additionally, the ability to accept foreign genes can vary depending on the genotype, which may explain the lack of regenerants in this study (Sabbadini et al., 2019).
Transformation of cocoa leaves mediated by Agrobacterium
The use of the vacuum pump method in the genetic transformation of cocoa clone IMC-67 with Agrobacterium tumefaciens resulted in a 92% response rate for callus formation in 50 young leaf explants, which were resistant to kanamycin selection (Figure 4A). In contrast, the genetic transformation by Agrobacterium tumefaciens without vacuum pump showed a low response rate of 18% (Figure 4B). The kanamycin-resistant calluses demonstrated their ability to proliferate in the selection medium (Figure 4B), while explants that did not exhibit proliferation capacity did not show any response (Figure 4C-D).
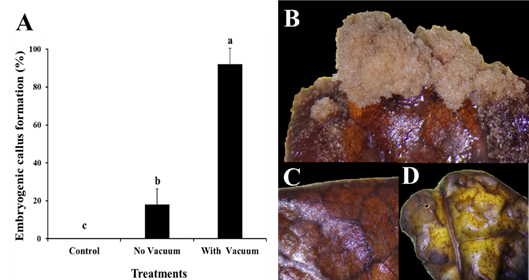
Figure 4 Genetic transformation of cocoa leaves clone IMC-67 with the AtBBM gene using the vector pk7WG2::AtBBM mediated by Agrobacterium tumefaciens. (A) Comparison of treatments in the formation of embryogenic calluses in cocoa leaves. (B) Calluses on live IMC-67 cocoa leaves in the sorting medium showing the formation capacity of embryogenic calluses. (C) Explants of cocoa leaves without response in the means of selection. (D) Explants controls of cocoa leaves IMC-67.
Recalcitrance of certain important fruit crops such as Theobroma cacao means a barrier in plant regeneration of plants using somatic embryogenesis, where the high composition of phenols inhibits this process. Furthermore, somatic embryogenesis is a genotype-dependent process that requires development and optimisation of new protocols to overcome this limitation (Pancaningtyas, 2015; Henao et al., 2018; Koné et al., 2019; Pérez-Caselles et al., 2021).
Embryogenic calluses were formed in IMC-67 cocoa leaves, which mainly presented a granular brown waxy appearance and allowed the observation of proembryogenic structures, as described by Henao et al. (2018). However, the formation of somatic embryos from these structures was not achieved. These findings are similar to those of Srinivasan et al. (2006), who used the vector 35S::BBM to study the effect of ectopic expression of AtBBM in tobacco and also failed to obtain somatic embryos but did observe callus formation and leaf wrinkling. It is important to note that embryogenic callus formation was achieved in a tissue that generally does not respond well to hormonal induction for callus formation or somatic embryogenesis, and the response is genotype dependent due to low expression of the BBM protein in this tissue. This study represents the first report on the development of genetic engineering applications through genetic transformation in Peruvian cocoa leaves clone IMC-67 (Florez et al., 2015; Fister et al., 2018). Furthermore, the calluses obtained can be used as useful explants for the production of secondary metabolites of interest, such as phenolic compounds (Rosabal et al., 2022).
Molecular analysis of cocoa transformed leaves
The PCR analysis results of 46 putative transgenic explants from the vacuum pump transformation process using AtBBM primers showed that all positive explants contained the AtBBM gene, as confirmed by the presence of an 1875 bp DNA amplicon (Figure 5). In contrast, only 9 positive explants containing the AtBBM gene were obtained from the agroinfiltration transformation process with Agrobacterium tumefaciens (Table 1). The use of AtBBM gene primers not only facilitated initial screening of transgenic plants but helped avoid false positives. Since the AtBBM gene was originally isolated from Arabidopsis thaliana, control plants also produced DNA bands when using specific primers of the AtBBM gene to detect gene integration. Specific primer combinations, such as front primers in the promoter and inverse primers in the gene, were used as a strategy to detect transgene integration.
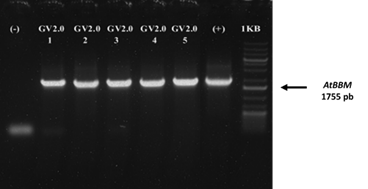
Figure 5 PCR analysis of cocoa transgenic embryogenic calluses of the IMC-67 clone with specific primers of the AtBBM gene. Positive plants contain the AtBBM gene (1755 bp).
Table 1 Transformation efficiency of IMC-67 cocoa leaves via formation of embryogenic calluses in two independent transformation treatments
| Treatment | No. of explants used | No. of explants with embryogenic calluses | PCR positives | Transformation efficiency (%) |
| Control | 50 | 0 | 0 | 0c |
| No vacuum | 50 | 9 | 9 | 18b |
| With vacuum | 50 | 46 | 46 | 92a |
Molecular analysis can rapidly detect transformation by examining the expression of the BBM gene in the true leaves of Theobroma cacao, as this TF has been shown to promote somatic embryogenesis (Jha & Kumar, 2018). Analyzing the response and functionality of the AtBBM gene accelerates response times and does not compromise genetic stability. This is particularly significant for studies on recalcitrant plants such as cocoa (Li et al., 2009; Lu et al., 2013; Guidarelli & Baraldi, 2015). Although the efficiency of the transformation did not lead us to produce somatic embryos or regenerated plants, but the response to the expression of the vector led to the production of embryogenic calluses, this result asserted a degree of participation of AtBBM in the leaf tissue of Theobroma cacao, obtaining a 92% efficiency of transformation using the infiltration to the vacuum, while by contact of solution with Agrobacterium tumefaciens transformed if the use of the vacuum pump was obtained an 18% efficiency of transformation, demonstrating that for the leaf tissue of Theobroma cacao is scarce transfection by contact due to certain unfavorable physicochemical conditions such as surface tension and viscosity, infiltrability characteristics and environmental factors that do not allow tissue infiltration (Chincinska, 2021).
4. Conclusions
The response of the transcriptional factor AtBBM in the formation of embryogenic calluses in cocoa leaves clone IMC-67 was evident in this study. Using the overexpression vector pk7WG2::AtBBM, a 92% response to the formation of embryogenic calluses in cocoa leaves was obtained with the vacuum infiltration method, mediated by Agrobacterium tumefaciens strain GV3101. Confirmation of the insertion of the overexpression vector in putative transgenic explants resulted in a transformation efficiency of 92%. This methodology allows for further studies of gene overexpression, insertion, silencing, and gene editing in Peruvian cocoa.