1. Introduction
Cannabis sativa is an annual plant belonging to the Cannabaceae family. It was botanically classified by Carl Linnaeus in 1753. The plant can grow up to 4 m tall, is dioecious, and has an erect stem with stipulate palmate leaves, the lower leaves are opposite, while the upper leaves are alternate (Ángeles López et al., 2014). The cannabis plant contains more than 566 compounds, including cannabinoids, alkanes, flavonoids, phenols, steroids, and terpenes (Fordjour et al., 2023; Gonçalves et al., 2019). Cannabinoids, such as THC and CBD, are used in the development of medicines to treat various diseases of the nervous system, neurodegenerative diseases, affective disorders, and for the modulation of eating behavior and have been used to treat rheumatic disorders (Montero-Oleas et al., 2020; Pauli et al., 2020; Russo, 2018). THC has psychoactive effects, whereas CBD does not, but has antipsychotic and anxiolytic effects (Boggs et al., 2018; Pintori et al., 2023).
Regarding the legal context of Cannabis sativa, Canada, Uruguay, and some states of the United States of America (USA) are among the countries that have completely legalized the cultivation, sale, possession, and consumption of Cannabis and its derivatives at the national level (Punja, 2021a). Colombia also has regulations that allow safe and informed access to the medical and scientific use of medical Cannabis, in addition to regulations associated with the licensing of the production and manufacture of cannabis derivatives and, finally a decree that allows the authorization of the use of dried cannabis flower for export for medicinal purposes (Departamento Administrativo de la FunciónPública, 2016;Ministerio de Salud y Protección Social, 2017,2021).
In recent years, interest in cannabis cultivation has increased due to its medicinal potential; however, this increase in larger-scale production has made the plant more susceptible to different contaminants, such as pesticides, mycotoxins, heavy metals, and microorganisms, which can have adverse effects on the physical, mental, behavioral and social health of consumers, especially in pediatric, immunocompromised, cancer and epileptic patients (Gwinn et al., 2023; Montero-Oleas et al., 2020; Vujanovic et al., 2020). Something that was evidenced by Dryburgh et al. (2018) who conducted a systematic review to assess the knowledge of cannabis contaminants and their impact on human health, identified that microorganisms, heavy metals, and pesticides are the primary contaminants in the harvest and postharvest stages and that there are also knowledge gaps in terms of health effects and interactions with the active components of Cannabis. There are reports of contamination with bacteria that have colonized cannabis cultivation, such as Escherichia coli, Salmonella spp., Acinetobacter spp., Bacillus spp., Burkholderia spp., Clostridium spp., Klebsiella spp., Pseudomonas aeruginosa, and Serratia spp.; regarding fungal con tamination, the most frequently reported are Asper gillus spp., Penicillium spp., and Fusarium spp. and also yeast genera such as Pseudozyma, Rhodotorula, Meyerozyma and Candida (Dryburgh et al., 2018; Punja et al., 2023; Vujanovic et al., 2020). Jerushalmi et al. (2020) conducted a study in which they identified leaf, stem, and soil pathogens affecting commercially grown medical Cannabis in Israel, finding that the most common pathogens are Alternaria alternata and Botrytis cinerea on leaves, Fusarium oxysporum and F. solani on stem and soil; additionally, fungi such as Aspergillus spp. and Penicillium spp. were isolated, which were considered in the study as a risk due to the possibility of mycotoxin formation.
Some of the species of the genus Aspergillus are noted for their ability to produce secondary metabolites hazardous to human and animal health such as mycotoxins, aflatoxins, ochratoxins, patulin, sterigmatocystin, and glyotoxin (Ámon et al., 2018; Malir et al., 2016; Moretti et al., 2017; Steenwyk et al., 2019) Some of the species of Aspergillus spp. that have been reported to produce ochratoxins are A. westerdijkae, A. carbonarius, A. niger, A. affinis, A. cretensis, A. flocculosus, A. muricatus, A. pulvericola, A. steynii, A. welwitschiae, A. subramanianii (Gil et al., 2020; Moretti et al., 2017).
Ochratoxins are toxic compounds that include ochratoxin A (OTA), which is the most dangerous due to the molecular structure that makes it difficult to degrade (Ding et al., 2023; Malir et al., 2016). OTA can cause negative health effects in humans and animals, both in the short and long term, including weight loss, dehydration, digestive hemorrhages, endemic nephropathy, carcinogenesis, and immunosuppression and is also considered a possible human carcinogen (group 2B) by the International Agency for Research on Cancer (IARC) (International Agency for Research on Cancer, 2012; Malir et al., 2016).
The presence of OTA in Cannabis is currently regulated in 26 USA jurisdictions, while the presence of Aspergillus spp. is held in 14 jurisdictions in the USA (Jameson et al., 2022). A study by Buchicchio et al. (2022) evaluated the presence of OTA in 142 cannabis samples, finding the presence of OTA in 27% of the samples ranging from 1.02 to 16.21 µg/kg. The stability of OTA in extracts derived from cannabis flowers has also been evidenced, establishing that after adding OTA to dried flowers, it remains and can be found in higher concentrations in the final extracts (Serafimovska et al., 2021). These facts highlight that the presence of OTA and Aspergillus spp. in medical cannabis inflorescences may represent a risk for the consumer. Therefore, this study aims to evaluate the contamination of medical cannabis inflorescences with ochratoxin-producing fungi and to determine the potential risk of synthesis of these mycotoxins by the associated fungal species.
2. Methodology
2.1 Collection of plant material
The collection of Cannabis sativa inflorescences was carried out at the Cubikan Group company in greenhouse crops, which is in Ansermanuevo, Valle del Cauca, Colombia, with an average temperature of 22.8 °C, an altitude of 1030 masl, average annual rainfall of 1216 mm, relative humidity of 60%, with a latitude of 4.7457177, and a longitude of -76.0480080.
Two commercial genotypes of medicinal Cannabis (F3IP and F8IP) from the Cubikan Group company were evaluated. For the isolation and in vitro identification of fungi present in the inflorescences, samples of medicinal Cannabis were collected between December 2020 and July 2021, ten samples were taken from each genotype, each weighing approximately 5 g; the samples were obtained after the drying process when the flower had a humidity between 12% and 14%.
The samples (Figure 1) were collected in sterile polyethylene bags and transported in a cooler with dry ice to the Phytopathology laboratory of the Department of Agricultural Production of the Faculty of Agricultural Sciences of the University of Caldas, located in the city of Manizales (Colombia), following the protocols established by the legislation (Ministerio de Salud y Protección Social, 2021).
2.2 Isolation of fungi present on postharvest inflorescences of medicinal Cannabis
To perform the isolation of fungi present in the samples, two techniques were performed, the inflorescences were placed directly on PDA agar (Criterion®), and in the other the inflorescences were disinfested using 70% alcohol for 30 s, followed by 1% sodium hypochlorite for 1 min, then three washes with distilled water each for 1 min, finally they were deposited on sterile paper towels to remove excess moisture.
Subsequently, for each of the techniques used, four inflorescences per Petri dish (0.8 g of wet flower) were placed on PDA agar (Criterion®) ensuring a good distribution. Thirty replicates were used for a total of 120 inflorescences per genotype. The isolates were incubated at 23 ± 2 °C (Thermo Scientific), simulating storage conditions. Once mycelial growth was observed around the inflorescences, part of this mycelium was transferred to new Petri dishes with PDA agar, which were incubated under the same temperature conditions to obtain pure cultures for identification, the evaluations were carried out every 24 h for ten days.
2.3 Taxonomic Identification
The fungi isolated from pure cultures were identified based on macroscopic and microscopic characteristics. Macroscopic characteristics were described using parameters such as colony margin, aspect, tonality of colonies, sporulation pattern, and color of conidial mass. Microscopic properties were observed using lactophenol blue under a light microscope (Leica DM500) with a 40X objective, as well as environmental scanning electron microscopy (ESEM) to study the morphology of fungal agents associated with cannabis inflorescences, in accordance with taxonomic descriptions found in the literature (Barnett & Hunter, 1998; Castaño, 2015; Castaño Zapata & Salazar Pineda, 1998). Subsequently, fungi with morphological characteristics compatible with the genus Aspergillus were stored for subsequent molecular identification.
2.4 Molecular identification
Once the purified isolates were obtained, total DNA was extracted using the DNeasy PowerLyzer PowerSoil Kit (Qiagen, Germany), according to the manufacturer's instructions. The product obtained was visualized on 1% agarose gel and quantified with a nanodrop (Thermo Scientific®). The final DNA concentration was adjusted to 15 ng/µl. The ITS ribosomal intergenic region was amplified to confirm the identity of the fungal genera identified by morphological methods (Table 1). The amplification conditions for the ITS region were as follows: initial denaturation at 95 °C for 1 min, followed by 94 °C for 30 s, 50 °C for 45 s, 72 °C for 1 min with 35 cycles, with a final extension to 72 °C for 10 min.
For the identification of Aspergillus spp., the coding region of the calmodulin (CaM) gene was used (Villa et al., 2020) (Table 1). The amplification conditions for coding genes were 95 °C for 3 min, 95 °C for 30 s, 55 °C for 35 s, and 72 °C for 1 min, with 38 cycles, with a final extension of 72 °C for 10 min.
Table 1 Primers used for the identification of Aspergillus spp. present in samples of Cannabis sativa inflorescences at the postharvest stage

The PCR product was subjected to a two-strands capillary sequencing reaction (SANGER REF) performed by Macrogen Incorporated (Seoul, Korea). The resulting sequences were compared with ITS and CaM sequences from the GenBank database of the National Center for Biotechnology Information (NCBI). As for the confirmation of species identity, a BLAST was conducted, and the identification of the BLAST hits with 98% identity was accepted. For each gene or region, the corresponding alignments were performed using the CLASSIFIER algorithm with the database of the Ribosomal Database Project: Clustal Omega program (Wang et al., 2007). The resulting sequences were included in a concatenated phylogenetic analysis using Maximum likelihood estimation using the software IQtree (Nguyen et al., 2015). The stability of phylogenetic relationships was assessed using the bootstrap method based on 1000 replicates. The outgroup used was Hamigera avellanea. In addition to molecular identification, whole genome sequencing was performed.
2.5 Sequencing, libraries, data analysis, and assembly
The sequencing of the WGS (Whole genome shotgun) genomic DNA of the Aspergillus strain was performed using Truseq Nano DNA libraries (350) and the Illumina NovaSeq platform (South Korea), which generated paired reads of 150 bases. Assembly, annotation, and integrity/quality assessment of the Aspergillus strain genomes were performed with the SPADES assembly tool version 3.15.4 (Prjibelski et al., 2020).
2.6 Phylogenetic, gene analysis and data analysis of Aspergillus associated with Ochratoxin production
A phylogenetic study was performed using 87 genomes of species of the genus Aspergillus deposited in public databases. The phylogenetic tree reconstruction was performed using the sequence of 100 conserved single-copy proteins encoded in each genome analyzed. The analysis was performed by Maximum likelihood estimation using the software IQtree (Nguyen et al., 2015). The stability of phylogenetic relationships was assessed using the bootstrap method based on 100 replicates. The outgroup used was Penicillium flavigenum. In addition, the search for gene clusters associated with secondary metabolite and toxin production was performed with the antiSMASH tool (fungal version), the most widely used tool to detect and characterize biosynthetic gene clusters (BGCs) in fungi (Blin et al., 2021). Manual curation for toxin-associated genes was also performed using the Artemis program (Carver et al., 2012).
3. Results and discussion
3.1 Taxonomic and molecular characterization of Aspergillus spp.
In the taxonomic characterization, one isolate of Aspergillus spp. was identified at the genus level, based on its anamorphic state. The macroscopic characteristics of this genus exhibited white colonies with yellow dots, powdery texture, radial grooves in some cases, and a yellow reverse (Figure 2A and 2B). Microscopically, hyaline conidia and terminal vesicles supporting the phialides were observed (Figure 2C), which are in accordance with the literature description for the genus Aspergillus (Estrada & Ramirez, 2019; Pitt & Hocking, 2009). Using electron microscopy, conidia and terminal vesicles supporting the phialides characteristic of the genus Aspergillus were also observed (Figure 2D).
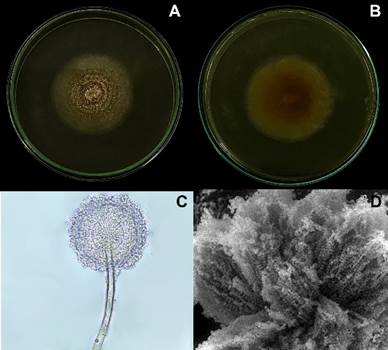
Figure 2 Macroscopic and microscopic features of Aspergillus westerdijkiae. A. Mycelial growth of A. westerdijkiae on the obverse, B. On the reverse, C. Microscopic features of A. westerdijkiae in light microscopy (40x). D. Microscopic features of A. westerdijkiae in scanning electron microscopy.
It has been documented that cannabis plants and cannabis-derived products can become contaminated with pathogenic microorganisms during various stages of production, ranging from cultivation to processing; this can pose an increased health risk, particularly for immunocompromised individuals who may inhale these contaminants. Seltenrich (2019) was one of the first authors to bring attention to the risk of Cannabis contamination with Aspergillus spp., a common indoor and outdoor fungus, where at that time, four species had been reported in the product that could cause infections (A. fumigatus, A. flavus, A. niger, and A. terreus). In a study by Jerushalmi et al. (2020), the main pathogens affecting the commercial cultivation of medical Cannabis in Israel were identified. Among the most common were Alternaria alternata, Botrytis cinerea, Fusarium oxysporum, F. solani, and Penicillium spp., as well as several species of Aspergillus, including Aspergillus flavus, Aspergillus fumigatus, Aspergillus niger, and Aspergillus westerdijkiae, known for their ability to produce some mycotoxins. On the other hand, Punja (2021b) studied the presence and fungal diversity in dried Cannabis inflorescences, identifying 34 different genera, among which the most relevant were Penicillium spp. Cladosporium spp., Botrytis spp., Talaromyces spp. and Alternaria spp. Likewise, some Aspergillus species such as A. flavus, A. niger, and A. ochraceus were found, also known for their risk of aflatoxin and ochratoxin formation. These studies show that contamination with Aspergillus spp. is frequent and is associated with both the harvest and postharvest stages.
3.2 Molecular Identification
Once purified fungal cultures were obtained in Petri dishes and identified using conventional methods, genus-level identification was performed using PCR of the ITS region of ribosomal DNA, and for molecular identification to determine the species within the Aspergillus genus, the CaM gene was used (Table 2). Overall, the analysis of the amplified product of the ITS region by PCR showed a band of approximately 560 bp, and for Aspergillus spp. using the amplified product of the CaM gene by PCR showed a band of 700 bp. When comparing the DNA sequences obtained from the ITS region with sequences from the NCBI GenBank using the BLAST tool, a first hit was obtained with a similarity percentage of over 95%. When comparing the sequences of CaM genes, combined with ITS sequence data, to those published in the NCBI GenBank, it was observed that the samples shared a 100% identity percentage with the nucleotide sequence of Aspergillus westerdijkiae species (Table 2).
Table 2 Classification of the Aspergillus genus according to the NCBI GenBank
| Region or gene | Genus | Species | Percentage of identity |
|---|---|---|---|
| CaM | Aspergillus | westerdijkiae | 99.4% |
The Maximum Likelihood phylogenetic analysis of the genetic region of Calmodulin (CaM), and the internal transcribed spacers of nuclear ribosomal DNA (ITS), revealed that genera morphologically identified as Aspergillus were closest to A. westerdijkiae species with a bootstrap support of 100% (Figure 3).
The Aspergillus spp. strain was sequenced and the complete genome of A. westerdijkiae was assembled using the Illumina NovaSeq platform. Finally, the genome was analyzed to identify its phylogenetic location and determine the genes that may encode for the biosynthesis of secondary metabolites. The genome had a size of 36.8 Mb, a GC content of 49.15%, and an N50 of 355.+055, comparable with the accession numbers LKBE00000000; LKBE00000000, MG701899, BioProject PRJNA679 179; reported in the literature (Chakrabortti et al., 2016; Gil et al., 2018; Han et al., 2016) (Table 3).
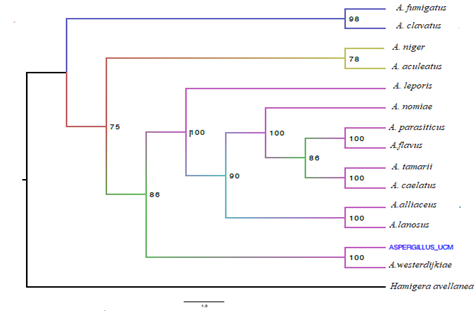
Figure 3 Concatenated Phylogenetic Tree (Maximum Likelihood Estimation) of the Ribosomal ITS Intergenic Region of Aspergillus and the CaM Coding Gene.
In the phylogenetic analysis, the for mation of a monophyletic clade with Aspergillus westerdijkiae was observed in the genus Aspergillus, supported by bootstrap values of 100, and a close relationship with A. melleus (Figure 4). When con ducting the analysis with whole genome data, it provides higher resolution power as it utilizes a larger set of genes, allowing for inference of phylo genetic relationships between A. westerdijkiae and other species within the Aspergillus genus (Han et al., 2016).
Table 3 Statistical Results of Aspergillus westerdijkiae genome assembly and Annotation
| Parameter | Value |
|---|---|
| Total length of sequence (bp) | 36890567 |
| Total number of sequences | 894 |
| Average contig length is | 41.264 |
| Largest contig (pb) | 886.915 |
| N50 stats | 355055 |
| % GC | 49.15 |
| % N | 1644 (0.00%) |
| Completeness (%C) | 97.0 |
| Complete and single copy (%S) | 96.7 |
| Complete and duplicated (%D) | 0.3 |
| Fragmented (%F) | 0.5 |
| Missing (%M) | 2.5 |
Several studies have demonstrated the presence of Aspergillus spp. as a common contaminant at harvest, in postharvest processes such as drying and in processed products such as resins and vapers (Pérez-Moreno et al., 2019; Punja, 2021b, 2021b; Punja et al., 2019; Sopovski et al., 2023). However, the presence of the fungus A. westerdijkiae, which was the species found in this study in dried inflorescence samples, has only been reported so far by Jerushalmi et al. (2020) on medicinal Cannabis leaves during the cultivation stage; these authors reported fungal contamination including A. flavus, A. fumigatus, A. niger, and A. westerdijkiae. The fungus A. westerdijkiae is known to contaminate coffee, grapes, cocoa, pepper, and some cured meat products (Amézqueta et al., 2012; Coton & Dantigny, 2019; Gallo et al., 2017; Paterson et al., 2014). It is also commonly associated with postharvest processes, such as drying or storage, and is characterized by its production of Ochratoxin A (Copetti et al., 2010; Gallo et al., 2017; Malir et al., 2016; Maman et al., 2021).
3.3 Analysis of genes associated of Aspergillus westerdijkiae with ochratoxin synthesis
The 70% of Aspergillus westerdijkiae strains have the ability to produce OTA, posing a threat to both humans and animals (Han et al., 2016; Sartori et al., 2014), for this reason genomic DNA sequencing of this species was performed to determine whether it had the genes coding for this mycotoxin.
The search for genomic regions that may encode non-ribosomal peptide synthetases (NRPS) was performed by bioinformatics analysis using the fungiSMASH platform, a tool that allows predicting possible gene clusters based on the presence of enzymes involved in different classes of biosynthetic pathways such as NRPS and polyketide synthases (PKS) (Chakraborty, 2022; Inglis et al., 2013).

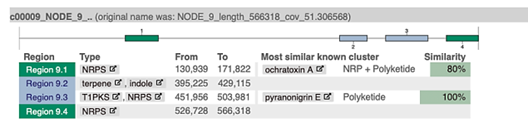
Figure 6 The cluster of genes encoding for ochratoxin biosynthesis in the Aspergillus westerdijkiae genome.
These analyses revealed that region 9.1 contained a group of biosynthetic genes that may eventually code for NRPS, involved in OTA synthesis with a similarity of 80% (Figure 5). The genome of A. westerdijkiae contains more than 50 secondary biosynthetic gene clusters, of which those with the highest expression are those encoding PKS and NRP, which is evidence that the strain found in this work has the capacity to synthesize secondary metabolites such as OTA (Chakrabortti et al., 2016; Schlösser & Prange, 2019). The complete gene cluster responsible for the synthesis of OTA in the metabolic pathway was manually curated and annotated and was found to be in the genome region of the genes under study (otaA, otaB, otaC, otaR, and otaD), with a higher occurrence of OTA genes observed (Figure 6). This finding is in agreement with previous reports by Susca et al. (2021) who mention that five genes (otaA, otaB, otaC, otaD, and otaR1) are directly involved in OTA biosynthesis. These five genes encode the following enzymes; halogenase (HAL), cytochrome P450 monooxygenase (p450), non-ribosomal peptide synthetase (NRPS), and polyketide synthase (PKS), in addition to a transcription factor bZIP, which regulates the expression of the rest of the genes (Susca et al., 2021).
IARC considers OTA a possible human carcinogen, classifying it in group 2B (International Agency for Research on Cancer, 2012). In addition, studies have documented several adverse health effects, such as hepatotoxicity, nephrotoxicity, immunotoxicity, teratogenicity, mutagenicity, genotoxicity, and embryotoxicity (Wang et al., 2022) Given that medical Cannabis is used in the treatment of various diseases of the nervous system, neurodegenerative and affective disorders, as well as to modulate eating behavior and treat rheumatic disorders, it is important to consider the risk that the possible synthesis of ochratoxins may generate for consumers, taking into account that this mycotoxin is difficult to degrade during processing stages (Liu et al., 2022; Montero-Oleas et al., 2020; Pauli et al., 2020; Russo, 2018)
To prevent OTA synthesis by A. westerdijkae and its contamination in cannabis, it is essential to adhere to recommendations such as implementing proper agricultural practices (GAP) during cultivation. Additionally, careful monitoring of environmental parameters during postharvest, including temperature, water activity (aw), pH, osmotic stress and the availability of carbon sources for the microorganism (Wang et al., 2023).
4. Conclusions
Through morphological and molecular analysis, Aspergillus westerdijkiae was identified in Cannabis sativa inflorescences. This fungal species is known for producing Ochratoxin A. In the present study, a cluster of genes encoding the biosynthesis of ochratoxins, including Ochratoxin A, was identified in the A. westerdijkiae strain found in medical cannabis inflorescences. This finding highlights a risk for the final consumer and underscores the need for medical cannabis producers to control environmental parameters, especially during postharvest, to prevent the production of this mycotoxin.